NYMU Taipei/My hot teams/Programmable cells
From 2007.igem.org
Contents |
Properties of Avian Influenza Viruses
Influenza viruses belong to the Orthomyxoviridae family. Antigenic differences in their nucleoprotein (NP) and matrix protein (M1) allow influenza viruses to be classified as types A, B or C (FIG. 1). For type A viruses, further subtyping is based on the antigenicity of the haemagglutinin (HA) and neuraminidase (NA) surface glycoproteins1. Currently, 16 HA (H1–H16) and 9 NA (N1–N9) subtypes have been identified in type A viruses. Type A influenza viruses have been isolated from various animals, including humans, pigs, horses, sea mammals and birds. Aquatic birds are thought to be the source of all influenza A viruses in other animal species. All 16 HA and 9 NA subtypes of type A viruses are maintained in aquatic bird populations, especially ducks, shorebirds and gulls.
Pathogenicity and Emergency
The emergence of high-pathogenicity avian influenza (HPAI) viruses in domestic poultry and the increasing number of cases of direct transmission of avian influenza viruses of different subtypes to humans are a significant threat to public health because of the potential for pandemic spread of these viruses. Research efforts to control emerging viral diseases are focused on improving surveillance and diagnostic methods, and on the development of antiviral drugs and effective vaccines. VACCINATION is the cornerstone of prevention, which will discuss in the next section. Interest in the development of pandemic influenza vaccines intensified with the outbreak of H5N1 influenza virus infections of humans in Hong Kong in 1997 and has increased further as H5N1 viruses have spread in birds and humans since 2003 (Subbarao and Joseph, 2007)
The Missing Link of Vaccine Design for Avian Viruses
Viruses could be antigenically distinguishable, but the consequences of these genetic and antigenic differences for vaccine development are not known. Are the antigenic differences that have been identified between avian influenza viruses of significance for human infections? Do viruses have similar pandemic potential? If the genetic differences between viruses are the result of co-evolution of an avian influenza virus with its avian host, and if the antigenic differences are driven by evolution in an avian host rather than by positive selection by an immune response to infection, then a vaccine generated against a virus one lineage might protect against a virus of another lineage. Unfortunately, this information is not known yet. Antigenic drift occurs when the genes encoding the HA and/or NA glycoproteins undergo stepwise mutations, resulting in variant viruses with amino-acid changes at one or more antibody-binding sites of HA and/or NA that allow the viruses to evade neutralization by antibodies generated as a result of previous natural infection or vaccination. However, the use of veterinary vaccines to protect poultry from infection with avian influenza viruses might drive evolution of the HA glycoprotein if such vaccines do not induce sterilizing immunity.
The viral determinants of pathogenicity of avian influenza viruses in humans are multigenic. Further studies are required to understand how the pathogenicity of avian influenza viruses affects the infectivity and transmissibility of these viruses in humans and to establish whether these factors have implications for vaccine design (Horimoto and Kawaoka, 2005; Subbarao and Joseph, 2007).
iGEM Goal
The increasing number of recent outbreaks of highly pathogenic avian influenza A virus infections (H5 and H7 subtypes) in poultry and in humans (through direct contact with infected birds) have raised concerns that a new influenza pandemic will occur in the near future, and underscores the need for control strategies to prevent an influenza pandemic.
The eradication of pathogenic avian influenza viruses seems to be the most effective way to prevent influenza pandemics; vaccination is the key strategy at present to prevent severe illness and death from pandemic influenza, although this strategy has not proven successful so far. Therefore, in our project, we wish to develop a system for mimicking the “natural evolution” to predict the viral antigenic mutation or “oncoming” new variant strain in a “short time” (directed evolution (Haseltine and Arnold, 2007)). According to these “new mutants”, which can reduce the possibility of low-sterilizing immunity of vaccines, further prevent the vaccine to drive evolution.
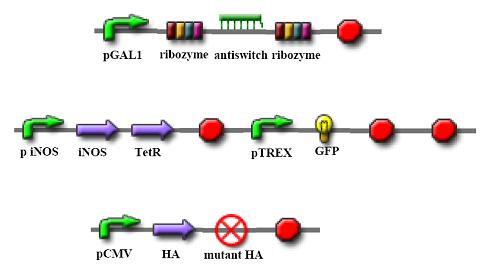
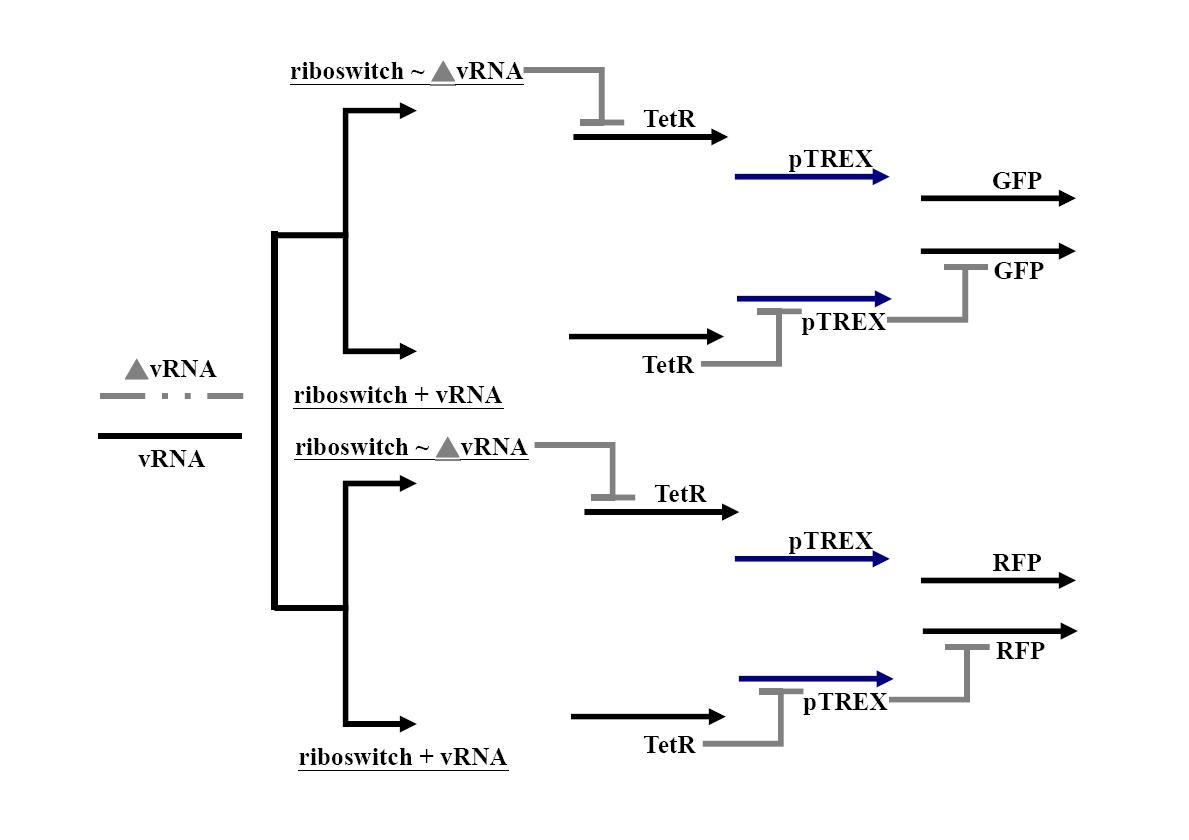
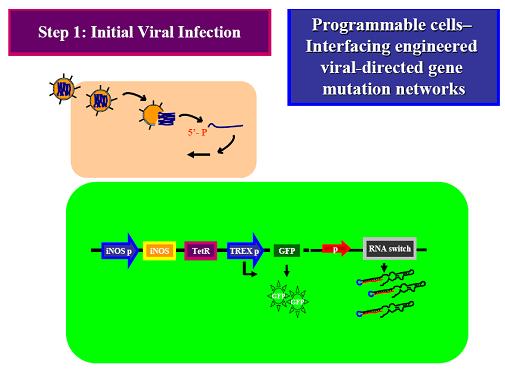
The goal of our project is to develop a “viral mutation biosensor”, which is capable extruding its arm upon adding a small ligand (theophyline). This viral mutation biosensor was existed in the “programmable cells”, which has the engineered gene networks for viral-directed mutation. The engineered networks consists a “viral mutant biosensor“and a system combined of accelerating mutation and reporter device. iNOS has demonstrated that accelerating viral mutation by nitration of guanine (Akaike et al., 2000; Akuta et al., 2006; Yoshitake et al., 2004), which can be incorporated into gene during viral replication. This accelerating mutation device can be naturally turned on by binding interferon (IFN) to iNOS promoter (Basler and Garcia-Sastre, 2007); the IFN releasing was induced by virus infection. We hope to drive viral evolution in this way rather then driving by un-designable vaccine. We hope that once this system is successful, it will be easy to refine it to be specific to recognize the new antigenic mutation, even the antigenic drift. To achieve this goal, we will use a riboswitch (Bayer and Smolke, 2005; Isaacs et al., 2006), which has a loop region to control the encoding of TetR (tetracycline regulator), a protein binding to the TREX promoter in front of GFP gene, that in the “off” state will inhibit the GFP production, while in the “on” state will allow GFP translation. This control would be happening at the mRNA level. The RNA probe can travel in the “programmable cell” and use a part of sequence we designed to target the viral sequence we interested (ex: HA receptor binding site versus new viral serotype) and sense the new mutant by loosing the hybridization ability of viral gene. Therefore, we can immediately sort the programmable cells containing mutant virus by green fluorescence and analysis these mutant antigenic type. A riboswitch is an RNA construct that resides in the 5` UTR of the gene it regulates. It contains an aptamer that will adopt different conformations in the presence or absence of a specific ligand. In our system, when the ligand (theophylline) is present, the riboswitch will extrude the anti-target region to hybridize the viral gene we designed, while not in the viral gene mutation, which loop region is then proceeding to bind to TetR mRNA and TetR translation blocks. After translation of TetR blocking, TREX promoter can normally activate GFP translation, which was the indicator of viral gene mutation.
The overall approach will be two-fold. The first aspect will be to start accelerating viral gene mutation by NO release induced by viral infection. The components are required to synthesize our system (in parallel) that the components interact properly and as expected. Secondly, it will be done to determine what gene differentiation (such as mutation or even antigenic drift), if any, are required to make get the best results from the system.
Thus our perspective goal is to expand the number of riboswitches containing different target gene we interested and to combine them with existing biobricks to create novel systems.
Parts and Device
Circuits
Illustration
Step 1: Initial Viral Infection (No IFN release) (green fluorescence- selection)
Step 2a: Viral genome mutation ( green fluorescence)
Step 2b: Viral genome no mutation (no green fluorescence)
References
Akaike, T., Fujii, S., Kato, A., Yoshitake, J., Miyamoto, Y., Sawa, T., Okamoto, S., Suga, M., Asakawa, M., Nagai, Y., and Maeda, H. (2000). Viral mutation accelerated by nitric oxide production during infection in vivo. Faseb J 14, 1447-1454.
Akuta, T., Zaki, M.H., Yoshitake, J., Okamoto, T., and Akaike, T. (2006). Nitrative stress through formation of 8-nitroguanosine: insights into microbial pathogenesis. Nitric Oxide 14, 101-108.
Basler, C.F., and Garcia-Sastre, A. (2007). Sensing RNA virus infections. Nat Chem Biol 3, 20-21.
Bayer, T.S., and Smolke, C.D. (2005). Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat Biotechnol 23, 337-343.
Haseltine, E.L., and Arnold, F.H. (2007). Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct 36, 1-19.
Isaacs, F.J., Dwyer, D.J., and Collins, J.J. (2006). RNA synthetic biology. Nat Biotechnol 24, 545-554.
Subbarao, K., and Joseph, T. (2007). Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol 7, 267-278.
Yoshitake, J., Akaike, T., Akuta, T., Tamura, F., Ogura, T., Esumi, H., and Maeda, H. (2004). Nitric oxide as an endogenous mutagen for Sendai virus without antiviral activity. J Virol 78, 8709-8719.