Paris/transclusion example
From 2007.igem.org
This is a test on the use of the transclusion directive. What you see below is the inclusion of text from other pages, as an example, the contents of the calendar for July.
Contents
|
July 2
yesterday -- tomorrow
First day in the lab !
Overview of the project
Planning of the lab work
Primer design
Preliminary work on w121 strain
Transduction of DapA- deletion from w121 to MG1655
We got the W121 strain from a lab in Pasteur Institute. This strain is [DapA-; Erythromycin R], but also has a couple of other mutations we are not interested in. Thus we will need to do a transduction of the deletion to the strain we will use: MG1655
We launched ON (= OverNight) culture of w121 to prepare a stock of P1 phages.
Measuring kinetic parameters of our strains
In order to model the dynamics of the synthetic organism, we need to measure several parameters. Two of which are:
- The growth of the [DapA-] strain relative to the DAP concentration of the medium;
- The excretion of DAP by the [DapA+] strain.
The first one is easy, we just need to measure growth kinetics of our [DapA-] strain contingent on the concentration of the DAP we add in the medium. The second one is more tricky. Direct dosage of DAP is quite complicated and expensive, thus we will try a kind of bio-measurement. We will grow the [DapA+] strain ON, then we will centrifugate to recover the culture medium. We will filter this culture medium to sterilize it and we will grow a [DapA-] strain in it. The growth curve should enable us evaluating the medium concentration in DAP.
July 3
Preparation of the P1 stock on the w121 strain.
See protocols. This preparation is necessary for transduction of DapA deletion in MG1655 strain.
Kinetic measurements:
As previously described, we want to measure :
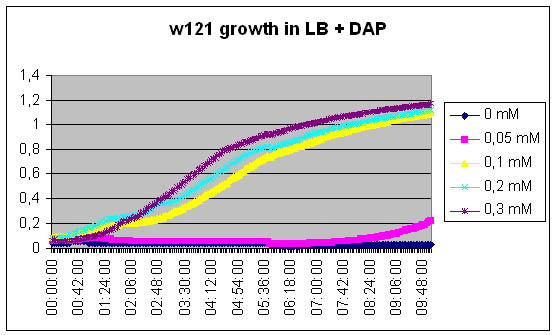
- Growth of DapA- strain (w121) relative to the concentration of DAP in the medium :
- The excretion of Dap by MG1655 DapA+ strain by an indirect way
Growth of DapA- strain (w121) relative to the concentration of DAP in the medium
We measure the kinetic of the w121 as a function of DAP concentration
[DAP] = 5mM
| Kinetic Array :Growthing of w121 strain 'supplemented' with S0 (1µL w121 strain incubated ON) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | ||||||||||||
| B | 200µL LB + 0µL DAP | 200µL LB + 2µL DAP | 200µL LB + 4µL DAP | 200µL LB + 8µL DAP | 200µL LB + 12µL DAP | |||||||
| C | ||||||||||||
| D | ||||||||||||
| E | ||||||||||||
| F | ||||||||||||
| G | ||||||||||||
| H | ||||||||||||
See Results.
Measuring excretion of Dap by MG1655 DapA+ strain in an indirect manner
We measure the growth of DapA- strain in the filtered growth medium of previously incubated MG1655 strain (S0).
S0 = Centrifuged and filtered medium of MG1655 ON
[DAP] = 5mM
| Kinetic Array :Growthing of w121 strain 'supplemented' with S0 (1µL w121 strain incubated ON) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | ||||||||||||
| B | 200µL S0 + 0µL DAP | 200µL S0 + 2µL DAP | 200µL S0 + 4µL DAP | 200µL S0 + 8µL DAP | 200µL S0 + 12µL DAP | |||||||
| C | ||||||||||||
| D | ||||||||||||
| E | ||||||||||||
| F | ||||||||||||
| G | ||||||||||||
| H | ||||||||||||
See Results.
Acinetobacter calcoaceticus ADP1
Culture
We received the strain Acinetobacter calcoaceticus ADP1 from Pasteur Institute (Center of biological resources of the Pasteur Institute): code CIP (ATCC number 33305). The strain was dehydrated and cultured following this protocol on LB agar plate at 30°C overnight.
See protocols
Medium to triglyceride/wax esters synthesis
Acinetobacter ADP1 is able to multiply in LB medium at 30°C. It is also able to synthesise lipids (triglyceride and wax ester) under specific conditions: Low Nitrogen Minimum Medium (LNMM).
This medium contains low nitrogen leading the bacterium to decrease its growing. With carbon hydrate source (here succinic acid), the bacteria stock energy into lipids (triglyceride and wax esters).
See protocols
<<home
July 4
Transduction to MG1655 using the P1 stock made on w121.
See protocols.
Growth kinetics : Results of July, 3
Growth of DapA- strain (w121) relative to the concentration of DAP in the medium
Measuring excretion of Dap by MG1655 DapA+ strain in an indirect manner
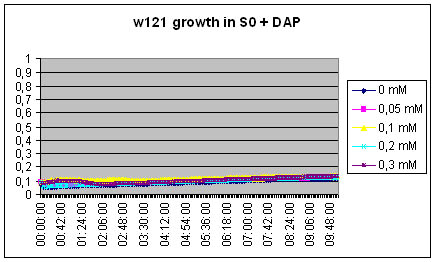
The w121 did not grow at all in the S0 medium... Either we did something wrong, or there is a logical explanation ! There might be growth inhibitors in the ON medium, their might be no nutriments left in the medium...
We will redo the experiment but this time, the medium will be recovered from exponential phase culture of MG1655
Acinetobacter liquid culture
We picked up a colony from LB solid culture from 07/03/2007 and put it into 5mL LB medium (30°C aerobic) (overnight culture).
Low Nitrogen Minimum Medium with Nile Red
This medium allows Acinetobacter ADP1 to synthesize triglyceride and wax ester. Lipid inclusions are seen by fluorescent dye (Nile Red).
See protocols.
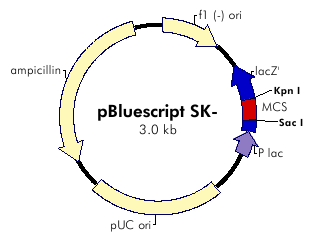
pKS::DGAT Plasmid: transformation
The plasmid contains ampicilline resistance gene and pLac promotor upstream the transgene DGAT (Diacylglycerol Acyltransferase).
We cloned the plasmid by bacterial transformation (Subcloning Efficiency DH5alpha Competent Cells: See protocols).
July 5
Preliminary work on w121 strain
As previously observed, the w121 strain did not grow at all in the S0 medium... We supposed that their might be no nutriments left in the medium because we used a medium in which MG1655 grew ON... We will redo the experiment but this time, the medium will be recovered from exponential phase culture of MG1655
- We started a culture 150 ml of MG1655, and we take samples of the culture at different OD during exponential phase :
- At OD = 0.2 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.2
- At OD = 0.4 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.4
- At OD = 0.6 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.6
- At OD = 0.8 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.8
Transduction of the DapA deletion into MG1655 failed...
We will redo the stock of P1 phages.
Glycerol stock of Acinetobacter ADP1
See protocols
Solid culture of Acinetobacter ADP1
We spread Acinetobacter on agar LNMM with and without Nile Red.
We spread E.Coli DH5alpha transformed by pKS::DGAT or not on agar LNMM with and without Nile Red.
See July6 for the results.
July 6
Preparing growth medium for transduction screening
If the transduction works (i.e. an homologous recombinaison that should delete DapA by inserting an erythromycin resistant cassette), transducted MG1655 Coli should grow in a medium LB+erythro+ DAP but can't grow in a medium LB+erythro without DAP.
- Preparation of DAP solution from the powder (50mM). See Protocols
- Making 10 petri dish (LB+tet+citrate+DAP). See Protocols
- Making 10 petri dish (LB+erythromycin+citrate+DAP). See Protocols
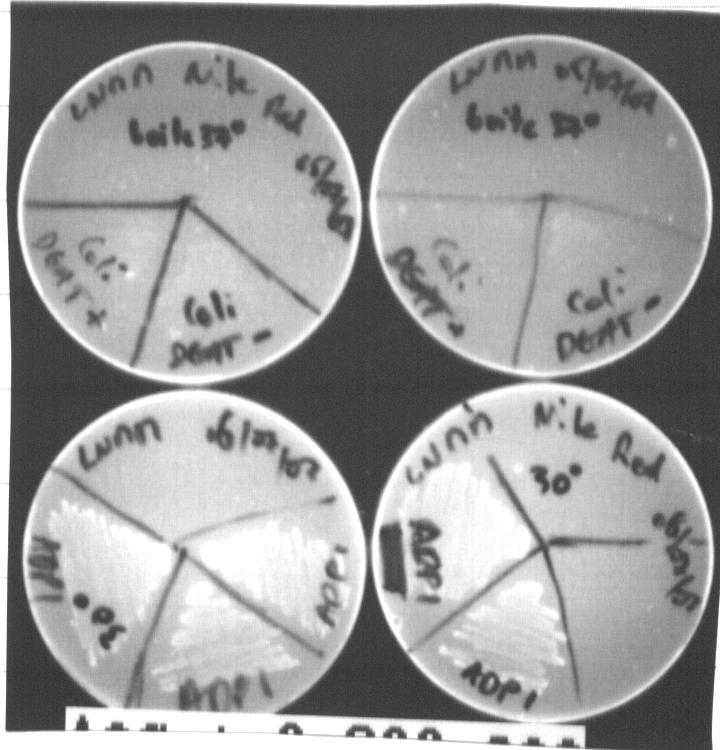
Acinetobacter and E.Coli on LNMM Nile Red solid culture
Nile Red can be excited by light around 312nm (Spiekermann,1999). After 24h incubation, we observed Acinetobacter and E.coli on LNMM with and without Nile Red:
We can observe basic fluorescence with Acinetobacter on LNMM with and without Nile Red. With E.coli transformed or not with pKS::DGAT, we do not observe fluorescence; indeed, coli do not grow on LNMM...
We decided to wait more to see acccumulation of triglyceride with Acinetobacter.
July 7
Preparing growth medium for transduction screening
If the transduction works (i.e. an homologous recombinaison that should delete DapA by inserting an erythromycin resistant cassette), transducted MG1655 Coli should grow in a medium LB+erythro+ DAP but can't grow in a medium LB+erythro without DAP.
- Preparation of DAP solution from the powder (50mM). See Protocols
- Making 10 petri dish (LB+tet+citrate+DAP). See Protocols
- Making 10 petri dish (LB+erythromycin+citrate+DAP). See Protocols
Acinetobacter and E.Coli on LNMM Nile Red solid culture
Nile Red can be excited by light around 312nm (Spiekermann,1999). After 24h incubation, we observed Acinetobacter and E.coli on LNMM with and without Nile Red:
We can observe basic fluorescence with Acinetobacter on LNMM with and without Nile Red. With E.coli transformed or not with pKS::DGAT, we do not observe fluorescence; indeed, coli do not grow on LNMM...
We decided to wait more to see acccumulation of triglyceride with Acinetobacter.
July 8
Preparing growth medium for transduction screening
If the transduction works (i.e. an homologous recombinaison that should delete DapA by inserting an erythromycin resistant cassette), transducted MG1655 Coli should grow in a medium LB+erythro+ DAP but can't grow in a medium LB+erythro without DAP.
- Preparation of DAP solution from the powder (50mM). See Protocols
- Making 10 petri dish (LB+tet+citrate+DAP). See Protocols
- Making 10 petri dish (LB+erythromycin+citrate+DAP). See Protocols
Acinetobacter and E.Coli on LNMM Nile Red solid culture
Nile Red can be excited by light around 312nm (Spiekermann,1999). After 24h incubation, we observed Acinetobacter and E.coli on LNMM with and without Nile Red:
We can observe basic fluorescence with Acinetobacter on LNMM with and without Nile Red. With E.coli transformed or not with pKS::DGAT, we do not observe fluorescence; indeed, coli do not grow on LNMM...
We decided to wait more to see acccumulation of triglyceride with Acinetobacter.