Saint Petersburg
From 2007.igem.org
(→Our Project) |
(→About Us) |
||
| Line 9: | Line 9: | ||
**Alexey Shalygin | **Alexey Shalygin | ||
**[https://2007.igem.org/User:Masha Maria Ditina] | **[https://2007.igem.org/User:Masha Maria Ditina] | ||
| - | ** | + | **[[Gennadiy Zakharov]] |
*'''Our Instructors:''' | *'''Our Instructors:''' | ||
| - | **Evgeny Zatulovskiy | + | **[[Evgeny Zatulovskiy]] |
**Vasiliy Romanov | **Vasiliy Romanov | ||
**[https://2007.igem.org/User:Alexej Alexej Skvortsov] | **[https://2007.igem.org/User:Alexej Alexej Skvortsov] | ||
Revision as of 09:36, 14 August 2007
Saint-Petersburg iGem2007 team
This year our team is participating for the first time in iGem2007 project.
Our team works at the [http://www.spbstu.ru Saint-Petersburg State Polytechnical University] in the department of Biophysics.
About Us
- Students:
- Tatiana Moiseeva
- Alexey Shalygin
- Maria Ditina
- Gennadiy Zakharov
- Our Instructors:
- Evgeny Zatulovskiy
- Vasiliy Romanov
- Alexej Skvortsov
Our Project
General
The aim of our project is to build on the E.Coli bacteria the copper sensor. To solve this problem we need to use the copper-homeostasis system of E.Coli. The most convenient for our purposes protein is CusR, the component of the Copper-Responsive Two-Component System.
We intend to use the CusR promoter with the reporter gene to build sensor.
So we perform a theoretical study of this problem.
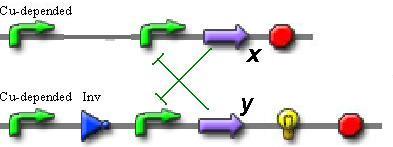
So, if we want to use only proteins without secondary metabolites, the best trigger sheme is the next:
In this scheme we can observe the switching between two repressors (x and y), depending on the transcription level from the copper-depended promoter.
But this scheme is rather difficult and need many constructing operations. So, we decided to think more simple scheme, using the secondary metabolites.