NYMU Taipei/ProjectDescription
From 2007.igem.org
(Difference between revisions)
(→Goal and system design) |
(→Mammalian Reporting System) |
||
| (23 intermediate revisions not shown) | |||
| Line 20: | Line 20: | ||
* Existed works (see References below) are focusing on how to take advantages from properties of mammalian cells to express and secrete insulin in a trail-and-error manner. However, from the perspective of synthetic biology, it is possible to engineer a cell works as we want, no matter what type of cells we apply to. | * Existed works (see References below) are focusing on how to take advantages from properties of mammalian cells to express and secrete insulin in a trail-and-error manner. However, from the perspective of synthetic biology, it is possible to engineer a cell works as we want, no matter what type of cells we apply to. | ||
---- | ---- | ||
| - | ==Goal and | + | ==Goal and System Design== |
* For the iGEM 2007 project, our team focuses on the development of a prokaryotic system to | * For the iGEM 2007 project, our team focuses on the development of a prokaryotic system to | ||
** express insulin regulated by external glucose concentration and | ** express insulin regulated by external glucose concentration and | ||
| Line 27: | Line 27: | ||
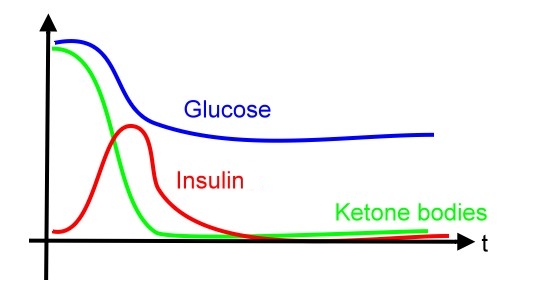
* system specification is shown below: | * system specification is shown below: | ||
[[Image:GAI-t curve.jpg|center|500px]] | [[Image:GAI-t curve.jpg|center|500px]] | ||
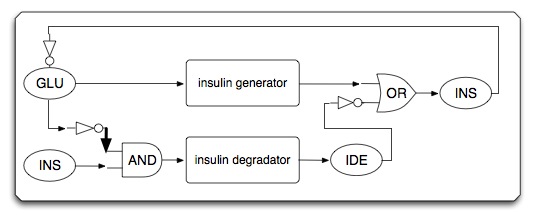
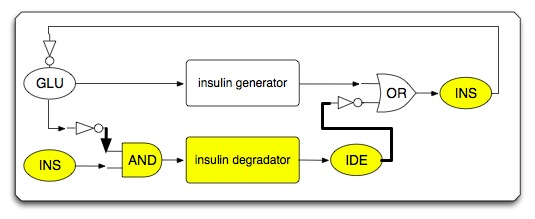
| - | * | + | * System diagram (initial state) |
[[Image:IGEM2007 insulin circuit diagram.png|center]] | [[Image:IGEM2007 insulin circuit diagram.png|center]] | ||
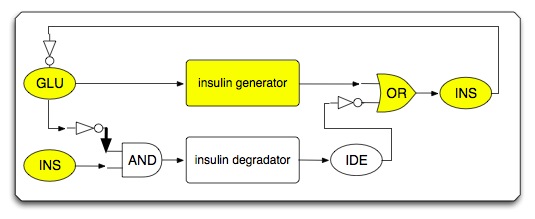
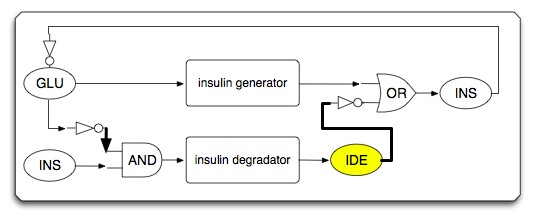
| - | * | + | * System under high glucose condition (insulin generating circuit is activated) |
[[Image:High_glucose.png|center]] | [[Image:High_glucose.png|center]] | ||
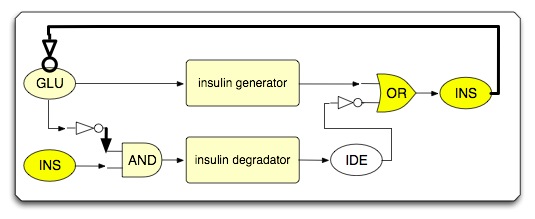
| - | * | + | * System under transition state (insulin increases glucose uptake; whole system shifts from insulin generation to insulin degradation) |
| - | [[Image: | + | [[Image:Trans_glucose.png|center]] |
| - | * | + | * System under low glucose condition (when glucose is low enough, the insulin generation turns off and insulin degradation turns on) |
[[Image:Low_glucose.png|center]] | [[Image:Low_glucose.png|center]] | ||
| - | * | + | * System after insulin degradation (both circuits turn off and wait IDE to be naturally degraded; back to initial state) |
| + | [[Image:Post_glucose.png|center]] | ||
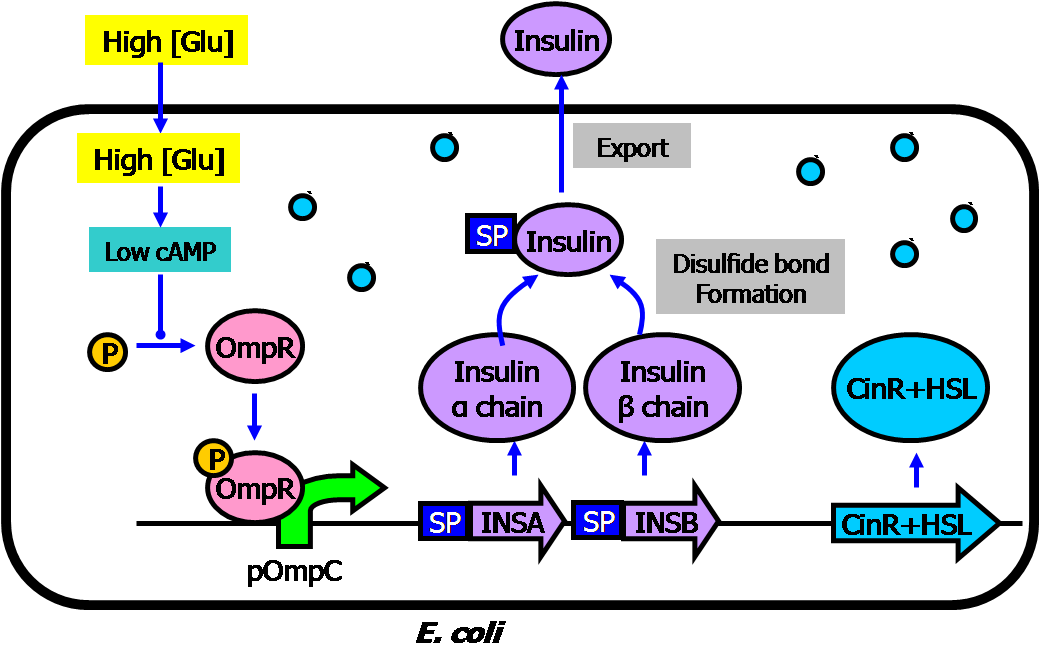
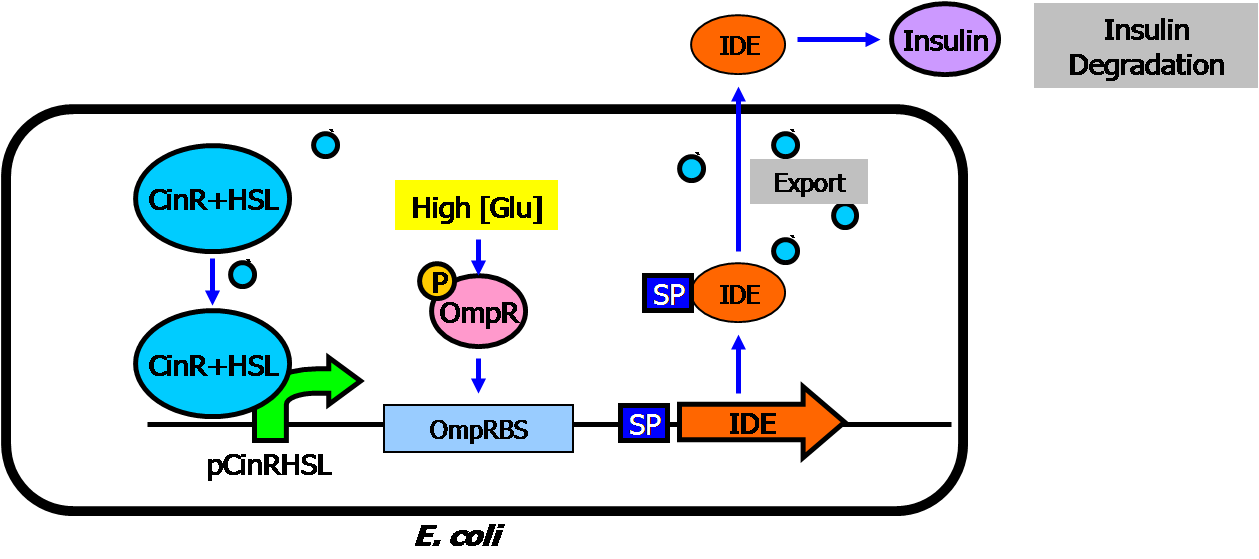
| + | * Circuit #1: high glucose-triggered insulin generator | ||
** The basic architecture for our insulin secretion design is shown in the figure below: | ** The basic architecture for our insulin secretion design is shown in the figure below: | ||
** External glucose can be sensed by GSD (Glucose Sensing Domain) from gene RcsC and transmits response to ERD (Responsive Domain from EnvZ gene) | ** External glucose can be sensed by GSD (Glucose Sensing Domain) from gene RcsC and transmits response to ERD (Responsive Domain from EnvZ gene) | ||
| Line 41: | Line 43: | ||
** With signal peptide (SP), the assembled insulin (by disulfide bonding) can be exported outside the membrane of the prokaryotic cells | ** With signal peptide (SP), the assembled insulin (by disulfide bonding) can be exported outside the membrane of the prokaryotic cells | ||
[[Image:Insulin basic2.png|center|500px]] | [[Image:Insulin basic2.png|center|500px]] | ||
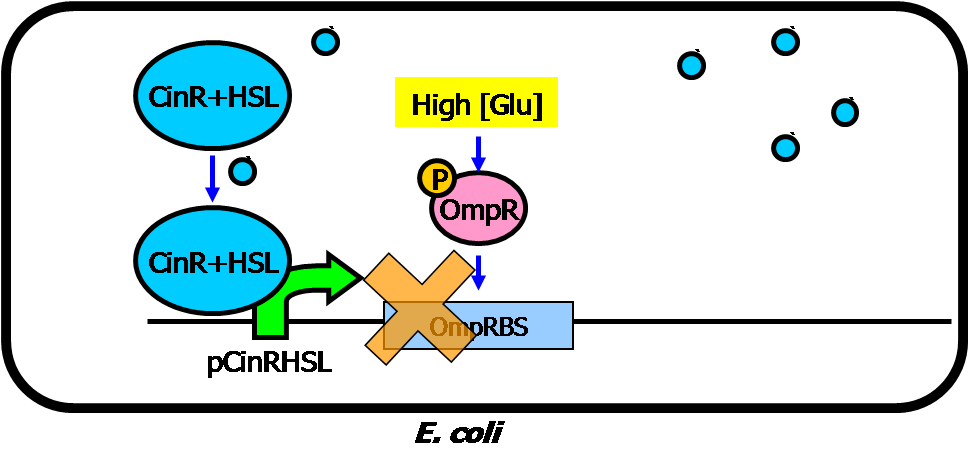
| - | * | + | * Circuit #2: low glucose and insulin triggered insulin inhibitor/destroyer |
[[Image:NYMU_Taipei_insulin_inhibitor_2.png|center|500px]] | [[Image:NYMU_Taipei_insulin_inhibitor_2.png|center|500px]] | ||
[[Image:NYMU_Taipei_insulin_inhibitor.png|center|700px]] | [[Image:NYMU_Taipei_insulin_inhibitor.png|center|700px]] | ||
| Line 51: | Line 53: | ||
* [[NYMU_Taipei/Protocols| Protocols]] | * [[NYMU_Taipei/Protocols| Protocols]] | ||
* [[NYMU_Taipei/Primer_List | Primer List]] | * [[NYMU_Taipei/Primer_List | Primer List]] | ||
| + | ---- | ||
| + | ==Verification== | ||
| + | ===pOmpC characterization by testing circuit #1 pOmpC + EYFP=== | ||
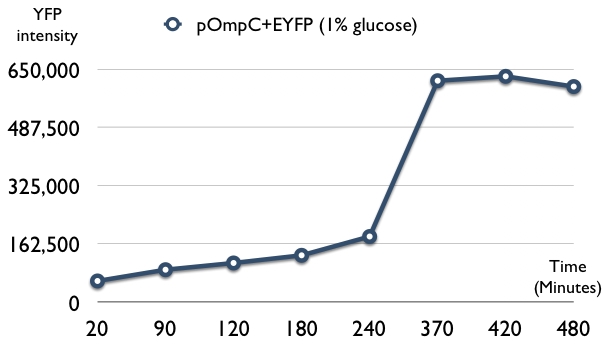
| + | * first trail: glucose can induce expression pOmpC operon when glucose concentration higher than 1% after 14h culture in the LB medium | ||
| + | [[Image:POmpC-EYFP test1.jpg|center]] | ||
| + | <br /> | ||
| + | * second trail: 1% glucose induction will activate the insulin expression after 4hr induction | ||
| + | [[Image:POmpC-EYFP test2.jpg|center]] | ||
| + | |||
| + | ===OmpRBS characterization by testing circuit #2 pLacIQ + OmpRBS + GFP=== | ||
| + | ===Mammalian Reporting System=== | ||
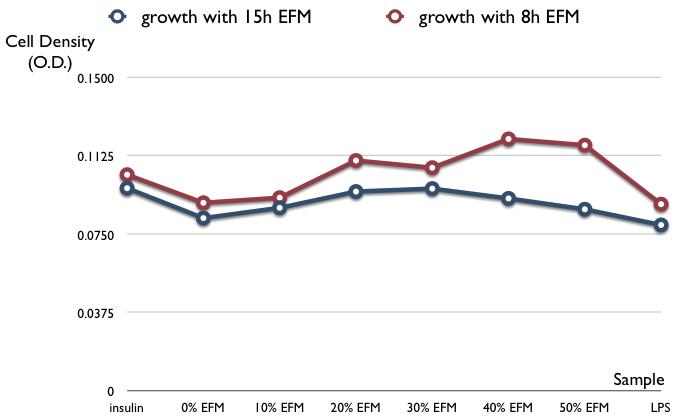
| + | * cellular growth effect to E.coli-free medium (EFM) after 15hr or 8 hr culture (12 replicates) | ||
| + | ** samples are as follows: no EFM, only insulin, 10-50% total volume of EFM, and LPS (toxic to cell) | ||
| + | [[Image:GWE_15h.jpg|center|500px]] | ||
| + | <br /> | ||
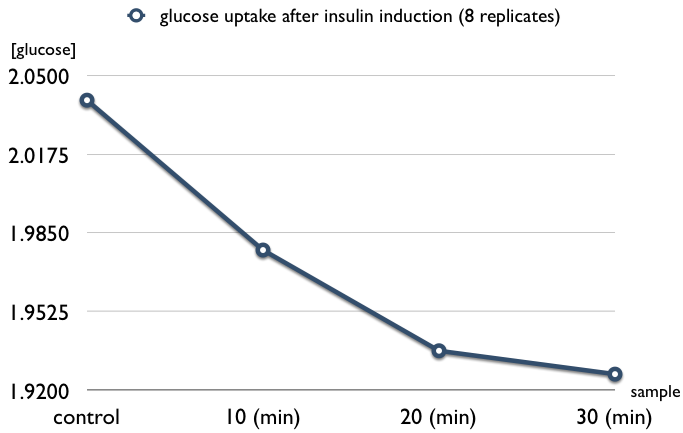
| + | * insulin induces glucose uptake in L6 cell (8 replicates) | ||
| + | ** samples are as follows: induced after 10, 20, and 30 minutes | ||
| + | [[Image:L6_INS_10-30.jpg|center|500px]] | ||
---- | ---- | ||
Latest revision as of 10:22, 27 October 2007
Project Description /
Lab Notes /
Part List /
Primer List
Contents |
GlucOperon
- A Glucose-based synthetic Operon system
- An automatic system in E.coli that operates blood sugar wonderfully!
- Insulin secretion system that promotes quality of life for diabetic patients
Summary
- Diabetes mellitus is a significant problem especially in developed countries and leads to several severe long-term complications. Compared to well-known type 2 diabetes mellitus, manifested with different degrees of insulin resistance, type 1 diabetes mellitus is caused by insulin deficiency following destruction of the insulin-producing pancreatic beta cells. Controlling blood sugar in a reasonable level and avoiding severe emergency as diabetes ketoacidosis (DKA) are very important clinical issues. Thus, the NYMU_Taipei iGEM 2007 team is designing a biological system to sense environmental glucose concentration and decrease the level of glucose by releasing insulin. Besides, life-protection functions for removing toxic ketoacids produced during DKA and preventing hypoglycemia status will also be established. This system will be a convenient and safe design for those patients with diabetes mellitus, and further improve their quality of life by avoiding them from diabetes-related morbidity and mortality.
Motivation
- Over 7 percent of adults in the United States (US) are known to have diabetes mellitus. However, because of the associated microvascular and macrovascular disease, diabetes accounts for almost 14 percent of US health care expenditures, at least one-half of which are related to complications such as myocardial infarction, stroke, end-stage renal disease, retinopathy, and foot ulcers. Compared to well-known type 2 diabetes mellitus, manifested with different degrees of insulin resistance, type 1 diabetes mellitus, one of the most common chronic diseases in childhood, is caused by insulin deficiency following destruction of the insulin-producing pancreatic beta cells. Data from the Center of Disease Control (CDC) show that more than 13,000 children and adolescents (19 years of age) are diagnosed with type 1 disease each year with a prevalence of 1.7 per 1000. Control of blood sugar and prevention of severe complications, such;as diabetic ketoacidosis (DKA), are important issues nowadays.
- Diabetic ketoacidosis (DKA) and hyperosmolar hyperglycemic state (HHS, also known as nonketotic hyperglycemia) are two of the most serious acute complications of diabetes. They are part of the spectrum of hyperglycemia and each represents an extreme in the spectrum. DKA is characterized by the triad of hyperglycemia, anion gap metabolic acidosis, and ketonemia. Metabolic acidosis is often the major finding. The serum glucose concentration is usually greater than 500 mg/dL (27.8 mmol/L) and less than 800 mg/dL (44.4 mmol/L). Three ketone bodies are produced in DKA: two ketoacids (beta-hydroxybutyric acid and acetoacetic acid), and one neutral ketone (acetone). The treatment of DKA and HHS is similar, including the administration of insulin and correction of the fluid and electrolyte abnormalities that may be present, including hyperglycemia and hyperosmolality, hypovolemia, metabolic acidosis (in DKA), and potassium depletion.
- Existed works (see References below) are focusing on how to take advantages from properties of mammalian cells to express and secrete insulin in a trail-and-error manner. However, from the perspective of synthetic biology, it is possible to engineer a cell works as we want, no matter what type of cells we apply to.
Goal and System Design
- For the iGEM 2007 project, our team focuses on the development of a prokaryotic system to
- express insulin regulated by external glucose concentration and
- clean the ketone bodies accumulated by mamalian cells due to insufficient intake of glucose
- balance between glucose and insulin to stablize glucose level and prevent low blood glucose condition
- system specification is shown below:
- System diagram (initial state)
- System under high glucose condition (insulin generating circuit is activated)
- System under transition state (insulin increases glucose uptake; whole system shifts from insulin generation to insulin degradation)
- System under low glucose condition (when glucose is low enough, the insulin generation turns off and insulin degradation turns on)
- System after insulin degradation (both circuits turn off and wait IDE to be naturally degraded; back to initial state)
- Circuit #1: high glucose-triggered insulin generator
- The basic architecture for our insulin secretion design is shown in the figure below:
- External glucose can be sensed by GSD (Glucose Sensing Domain) from gene RcsC and transmits response to ERD (Responsive Domain from EnvZ gene)
- TCS phosphorylation relay activates the expression of insulin (both alpha and beta chains)
- With signal peptide (SP), the assembled insulin (by disulfide bonding) can be exported outside the membrane of the prokaryotic cells
- Circuit #2: low glucose and insulin triggered insulin inhibitor/destroyer
Implementation
Verification
pOmpC characterization by testing circuit #1 pOmpC + EYFP
- first trail: glucose can induce expression pOmpC operon when glucose concentration higher than 1% after 14h culture in the LB medium
- second trail: 1% glucose induction will activate the insulin expression after 4hr induction
OmpRBS characterization by testing circuit #2 pLacIQ + OmpRBS + GFP
Mammalian Reporting System
- cellular growth effect to E.coli-free medium (EFM) after 15hr or 8 hr culture (12 replicates)
- samples are as follows: no EFM, only insulin, 10-50% total volume of EFM, and LPS (toxic to cell)
- insulin induces glucose uptake in L6 cell (8 replicates)
- samples are as follows: induced after 10, 20, and 30 minutes
References
- Engineered enteroendocrine cells secrete insulin in response to glucose and reverse hyperglycemia in diabetic mice, 2007 Mol Ther.
- Development of genetically engineered human intestinal cells for regulated insulin secretion using rAAV-mediated gene transfer, 2003 Biochem Biophys Res Commun
- http://www.utdol.com/utd/content/topic.do
- Recombinant DNA Technology in the Synthesis of Human Insulin
- Glucose-Dependent Insulin Release from Genetically Engineered K Cells, 2000 science
- [http://www.nature.com/gt/journal/v7/n21/abs/3301306a.html Glucose-stimulated and self-limiting insulin production by glucose 6-phosphatase promoter driven insulin expression in hepatoma cells, 2000 Gene Therapy]