Boston University/Restriction Choice
From 2007.igem.org
Restriction Enzyme Choice
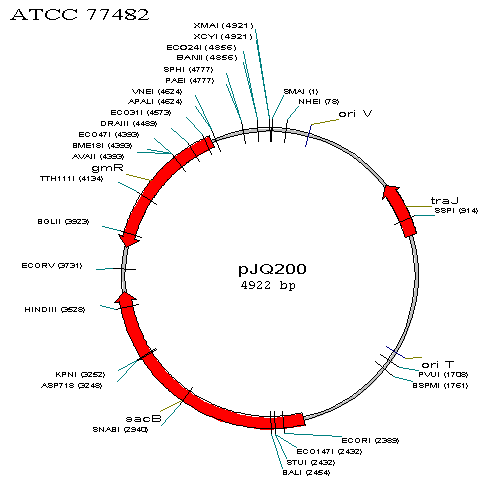
This is an image of PJQ200, not engineered.
We must engineer the plasmid in three ways:
- Remove the sacB gene so the plasmid will not be lethal in S. oneidensis if the bacteria is exposed to sucrose.
- Insert the mutated global transcription factors
- Allow for appropriate antibiotic selection.
We will combine the first 2 goals into one solution by removing the sacB gene and inserting in its place the transcription factors. We will use the restriction enzymes HindIII and EcoRI, whose sites bound the sacB gene.
In terms of antibiotic selection, after conjugation we will want to kill off all E. coli and those S. oneidensis that do not take up the plasmid, leaving only S. oneidensis with the plasmid. Ordinarily, selection by gentamicin will not be effective for our purposes because E. coli with an unaltered gmR plasmid will survive. In addition, because S. oneidensis normally is resistant to gentamicin, S. oneidensis even without the plasmid will survive. To solve this problem, we plan on removing the gtmR gene and replacing it with the kanR gene that gives resistance to kanamycin. Kanamycin will then kill off all E. coli without the plasmid and S. oneidensis without the plasmid. Then, in order to remove all E. coli with the plasmid, we can use gentamicin, as the plasmid will no longer give resistance to gentamicin. In order to remove gtmR and replace it with kanR, we will use restriction enzymes BsaI and Tth111I. BsaI is an isoschizomer with Eco31I, meaning BsaI will cut at the same place that Eco31I does. Eco31I and Tth111I's sites bound the gtmR gene.
Double Digests: Additional Food for Thought
submitted by Christian Ling
Another consideration we made when selecting pairs of restriction enzymes were their compatibility in solution: meaning, can the restriction enzymes be safely mixed together in a shared buffer in order to run a "double digest"? Although not absolutely necessary, performing double digests saves in time and effort as less buffers need to be made/used and the plasmid does not have to be repeatedly cleaned and then reinserted into different solutions of buffer/restriction enzymes. In order to aid us in this endeavor, we used the New England Biolab's Double Digest Calculator [http://www.neb.com/nebecomm/DoubleDigestCalculator.asp] to evaluate whether or not different combinations of restriction enzymes were conducive to double digestion.
Although incredibly easy to use, here are some basic instructions on using the Double Digest Calculator:
- Click on the Select 1st Enzyme Box. A drop-down menu should appear from which you can select your enzyme. Keep in mind that you can type in the enzyme name after initially clicking the box in order to speed things up.
- Click on the Select 2nd Enzyme Box and select your enzyme.
- Hit the GO Button to receive an evaluation of double digestion, as well as a mini-protocol for double digestion if it is possible.
Keep in mind are that it is not necessary to have 100% activity for the double digestion to work. Obviously 100% activity for both enzymes in the shared buffer will result in better yields, but our team has been reassured by grad students in the Gardner Lab that 50% activity and above will suffice.