Boston University/Zymo Transformation Results
From 2007.igem.org
< Boston University(Difference between revisions)
(→Overview of Zymo Transformation Results) |
|||
| (16 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
==Introduction to Zymo Transformation and Testing== | ==Introduction to Zymo Transformation and Testing== | ||
| + | :During the weekend of 7/14/07, the BU iGEM team ran a test of Zymo transformation of plasmids pBAD18s, pBAD30, and pjQ200 into wildtype S. oneidensis. Plasmids pBAD18s and pBAD30 were recommended to us by Professor Gardner as possible vectors for mutated Shewy genes. pjQ200 was the plasmid our team had previously selected as a prime candidate for serving as a vector. In order to check (and in the case of pjQ200, double-check) that these plasmids are capable of direct transformation into Shewy, tests of Zymo transformation and electroporation were conducted. For both types of transformation, E. coli was used as a control to verify that our transformation procedures were conducted properly. The results are posted below in a series of images and a short analysis. Lastly, gentamycin plates were used to select for pjq200 transformants, while ampicillin plates were used to select for pBAD18s and pBAD30 transformants. | ||
| + | |||
| + | ==Results of Zymo Transformation== | ||
{| style="width:700pt;" border="1" | {| style="width:700pt;" border="1" | ||
| | | | ||
| Line 45: | Line 48: | ||
|} | |} | ||
| - | + | ==Overview of Zymo Transformation Results== | |
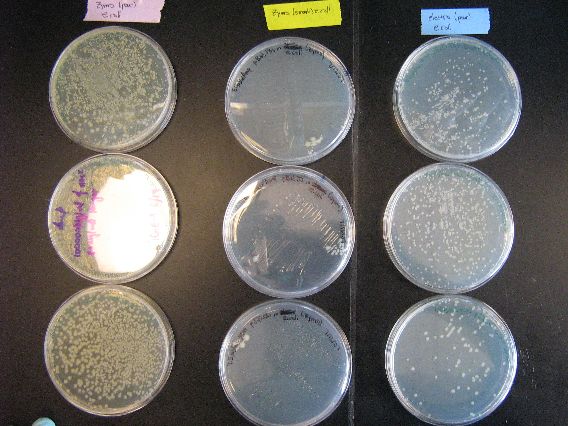
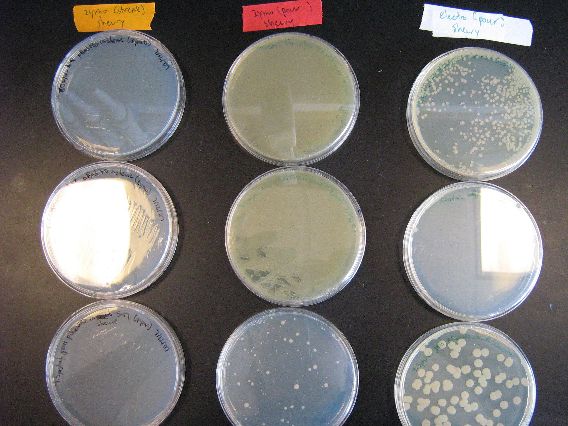
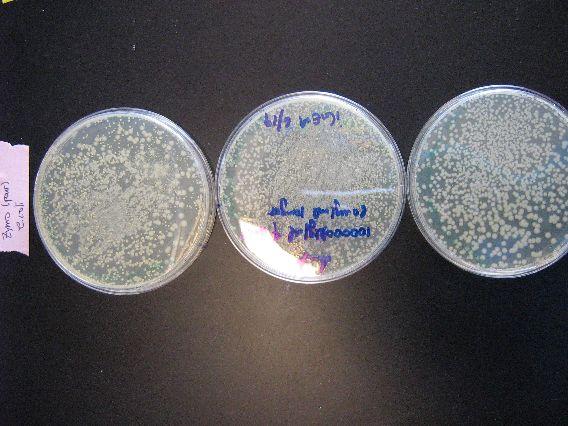
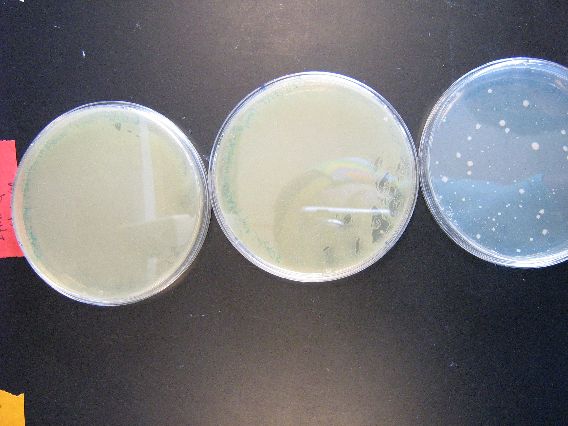
| - | + | :The results from the Zymo transformation went fairly well. Streaking of transformed E. coli (first picture, middle column) and Shewy (second picture, left column) resulted in minimal/no colonies forming, which suggests that the concentration of transformed cells is very low. However, the plates in which 100 microliters of transformed cells were poured had many colonies for E. coli (first picture, left column) and a thick lawn of Shewy (second picture, middle column). | |
| - | + | ||
| - | [[Image:BU_zymo_pour_ecoli.jpg]] | + | :There are some important things to note from these results. First of all, for the Zymo plates in which transformed Shewy was poured (second picture, middle column) the bottom plate has a scant few colonies as compared to the two lawn-covered plates above it. Since the top two plates in the column are pBAD18s and pBAD30, it appears that pjQ200 has a lower transformation efficiency into Shewanella than the other two plasmids. |
| - | + | ||
| - | [[Image:BU_zymo_pour_shewy.jpg]] | + | :Secondly, we believe that there is also fungal/yeast contamination for the two plates in which Zymo-transformed E. coli was poured (first picture, top left plate and the plate below it). Close examination reveals that there are two visually distinct types of colonies forming and there are peculiarly '''"sweet"''' and '''"delicious"''' smells coming from the plates, which suggests a yeast contamination. Coincidentally, the last time a set of results had a yeast contamination was when we were doing an earlier Zymo transformation with E. coli. We may need to check our Zymo kit for contamination. |
| - | [[Image:BU_zymo_streak_ecoli.jpg]] | + | |
| - | [[Image:BU_zymo_streak_shewy.jpg]] | + | |
| + | {|style="width:700pt;" border="1" | ||
| + | |[[Image:BU_zymo_pour_ecoli.jpg|left]] | ||
| + | | | ||
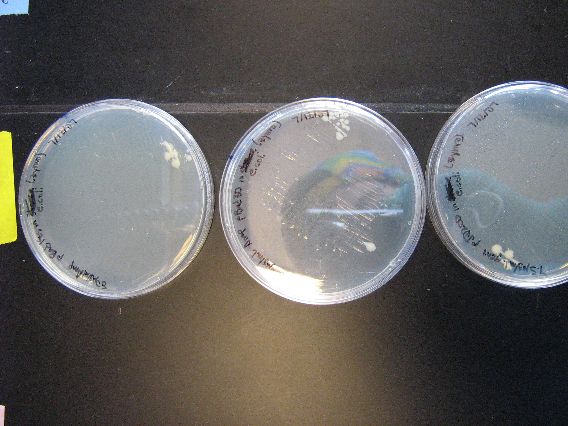
| + | :The plates to the left are the E. coli that we ran as controls for the Zymo transformation. More specifically, each plate had 100 microliters of transformed E. coli poured onto them. From left to right, the plasmid contents are as follows: pBAD18s, pBAD30, pjq200. | ||
| + | |||
| + | :As mentioned earlier in the discussion of the results, the plates containing plasmids pBAD18s and pBAD30 appear to have yeast contamination. Although the color of all the growths on these two plates appear to be similar, there are colonies on both plates that are very diffuse and "fuzzy" looking, and other colonies that are smaller, more granular, and yellow-tinged. We believe that the second, smaller colonies are yeast contamination. Thankfully, the rightmost plate appears to be (and smells like) a contamination free, E. coli only plate. | ||
| + | |- | ||
| + | |[[Image:BU_zymo_pour_shewy.jpg|left]] | ||
| + | | | ||
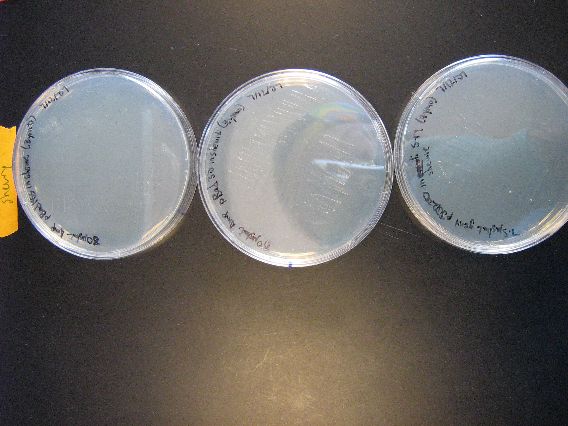
| + | :From left to right, the plasmid contents of the plates are pBAD18s, pBAD30, and pjq200. These plates had 100 microliters of transformed S. oneidensis poured onto them. | ||
| + | |||
| + | :The heavy lawns for the first two plates indicate that we definitely have successfully Zymo transformed S. oneidensis. However, the rightmost plate's lack of a lawn and smaller amount of colonies demonstrates pjq200's inferior transformation efficiency as compared to the pBAD plasmids. We may have to rethink our use of plasmid pjq200 if greater transformation efficiency is required. | ||
| + | |- | ||
| + | |[[Image:BU_zymo_streak_ecoli.jpg|left]] | ||
| + | | | ||
| + | :Once again, the contents of the plates from left to right are pBAD18s, pBAD30, and pjq200 all transformed into E. coli. The tiny patches of colonies visible on the edges of each plate demonstrate that the transformation efficiency of all our plasmids is a bit lacking. We will either need to streak more than 5 microliters of transformed cells in order to get more colonies, or somehow increase the transformation efficiency. | ||
| + | |- | ||
| + | |[[Image:BU_zymo_streak_shewy.jpg|left]] | ||
| + | | | ||
| + | :There was no growth at all for the plates in which 5 microliters of transformed Shewy were streaked onto antibiotic plates. As proven in this (and the last) set of plates, we will definitely need to plate large volumes of our transformed cells in order to harvest a suitable number of colonies to build our mutant library. | ||
| + | |||
| + | |} | ||
[[Boston University | Back]] | [[Boston University | Back]] | ||
Latest revision as of 20:54, 16 July 2007
Introduction to Zymo Transformation and Testing
- During the weekend of 7/14/07, the BU iGEM team ran a test of Zymo transformation of plasmids pBAD18s, pBAD30, and pjQ200 into wildtype S. oneidensis. Plasmids pBAD18s and pBAD30 were recommended to us by Professor Gardner as possible vectors for mutated Shewy genes. pjQ200 was the plasmid our team had previously selected as a prime candidate for serving as a vector. In order to check (and in the case of pjQ200, double-check) that these plasmids are capable of direct transformation into Shewy, tests of Zymo transformation and electroporation were conducted. For both types of transformation, E. coli was used as a control to verify that our transformation procedures were conducted properly. The results are posted below in a series of images and a short analysis. Lastly, gentamycin plates were used to select for pjq200 transformants, while ampicillin plates were used to select for pBAD18s and pBAD30 transformants.
Results of Zymo Transformation
Overview of Zymo Transformation Results
- The results from the Zymo transformation went fairly well. Streaking of transformed E. coli (first picture, middle column) and Shewy (second picture, left column) resulted in minimal/no colonies forming, which suggests that the concentration of transformed cells is very low. However, the plates in which 100 microliters of transformed cells were poured had many colonies for E. coli (first picture, left column) and a thick lawn of Shewy (second picture, middle column).
- There are some important things to note from these results. First of all, for the Zymo plates in which transformed Shewy was poured (second picture, middle column) the bottom plate has a scant few colonies as compared to the two lawn-covered plates above it. Since the top two plates in the column are pBAD18s and pBAD30, it appears that pjQ200 has a lower transformation efficiency into Shewanella than the other two plasmids.
- Secondly, we believe that there is also fungal/yeast contamination for the two plates in which Zymo-transformed E. coli was poured (first picture, top left plate and the plate below it). Close examination reveals that there are two visually distinct types of colonies forming and there are peculiarly "sweet" and "delicious" smells coming from the plates, which suggests a yeast contamination. Coincidentally, the last time a set of results had a yeast contamination was when we were doing an earlier Zymo transformation with E. coli. We may need to check our Zymo kit for contamination.
| |
| |
| |
|