Boston UniversityStatus
From 2007.igem.org
Contents |
What We've Accomplished
Week's (Ambitious) Goals
Week of 6/4:
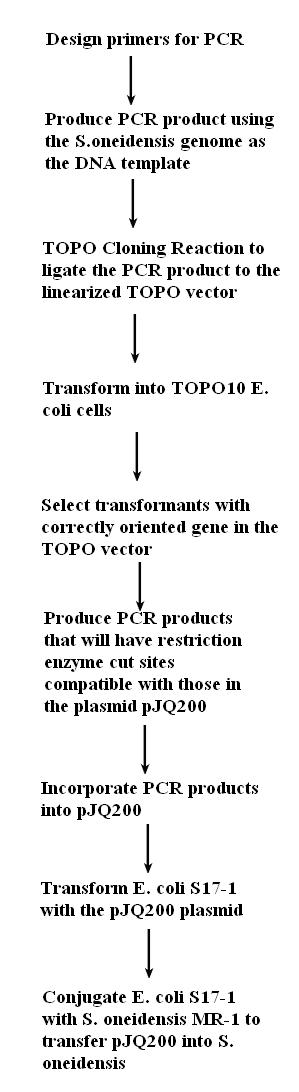
1. Evaluate the transformation that was done on Friday.
2. Confirm the correct plasmid (pJQ200)
3. Find appropriate restriction enzymes that cut by BLASTing all the plasmid's restriction enzyme sites onto the global transcription factors.
4. Re-design primers for the global transcription factors based on the restriction enzymes we have selected.
5. Order primers.
6. Practice regular (non-error prone) PCR with the primers to check that they work.
7. Incorporate the global transcription factors into the plasmid and transform this plasmid into the E.coli.
8. Conjugate this plasmid into Shewy.
Materials We Need
Error-Prone PCR: Need to Buy
Ligases: Need to Buy?
Short-Term To-Do List
Ordering of Error-Prone PCR Materials: Not completed
Thank-You Letters sent to Pfizer: Not Completed
Protocols
Calcium Chloride/Heat Shock Plasmid Transformations Protocol
pTrcHis TOPO TA Expression Kit Cloning Protocol
Question and Answer
Bold=new question/answer
Q: Can we get the kanR gene with appropriate sticky ends for BsaI and Tth111I? (DB)
A: yeah, we can get the DNA template for the kanR gene from the lab. we'll need to design some primers to PCR amplify it out with the BsaI and Tth111I cut sites. after cutting with the restriction enzymes, the kanR will have the appropriate sticky ends. frank suggested that we try ligating the kanR gene to the global regulator gene. (ie. kanR ligated to hlyU) and then stick that into the pJQ200. this way we can select for succesfull hlyU transformations with kanamycin
Q: Ok, good. Do those primers still need to be ordered? And if we follow Frank's suggestion, what will go in place of the gtmR gene?
A: the hlyU-kanR gene will be inserted into gmtR. the sacB will be left untouched if this method is used. the other method we can do is to just kill the gmtR gene, then insert hlyU into sacB. we will then add sucrose so any bacteria with a sacB site will die. since hlyU will be interrupting the sacB site, successfull transformations with hlyU will survive the sucrose selection
Q: So according to the first method, you'd have to never expose the bacteria to sucrose. Are we sure this is possible (some of the media might contain sucrose). According to the second method, won't bacteria without plasmids survive? I suppose we could expose them to more antibiotics then. Is there a problem with the plan I think I got out of our talks on Friday (ie the one on this wiki--inserting kanR into gtmR and hlyU into sacB?)
Q: Also, can someone update the primer design section? Also also, have thank you letters been sent?
A: i'll get on both of those by monday's end
Q: hey danny not quite sure how to make the image i just uploaded fit well (the figure with what we plan to do over the next month)
A: Looks pretty good to me
Q: Can I move the Lab Design and Notes sections to the What We've Accomplished section?
Relevant Publications and Links
https://www.atcc.org/common/catalog/numSearch/numResults.cfm?collection=mb-vector&atccNum=77482