Q-Antiterminator
Background Information
Q is a 207 amino acid protein with a mostly linear structure. It binds to lambda genomic DNA at Qut. The RNA polymerase is modified as it initializes transcription at pR', slightly downstream of Q. This modification causes the RNA polymerase to transcribe genes beyond the terminator tR'.
Role in Lambda Life Cycle
Since Q is located downstream of the terminator tR, its transcription requires the presence of N. During early stages of lambda's life cycle, as well as in the lysogenic pathway, the translation of Q is downregulated by an antisense RNA. The promoter pAQ transcribes Q in reverse, producing a Q-antisense RNA that binds to the Q messenger RNA and prevents ribosomal attachment. Furthermore, a relatively high concentration of anti-Q is required to effectively cause antitermination, giving the phage time to begin a lysogenic life cycle if conditions are appropriate.
Once Q has modified the RNA polymerase, the polymerase begins to transcribe genes beyond tR'. These genes are crucial to the production of new phage components, such as tails and heads (protein capsules). Once these genes are expressed, the phage is committed to the lytic pathway.
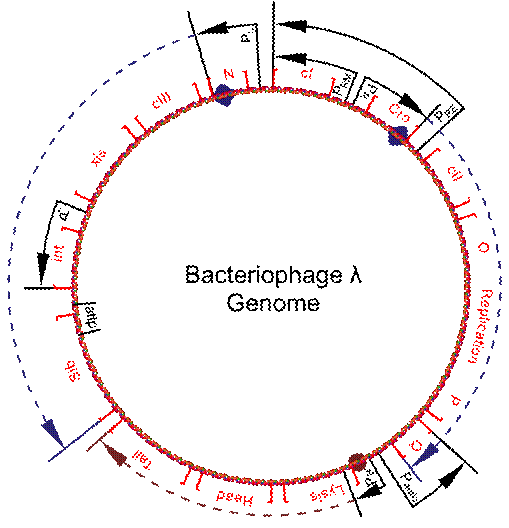 Schematic representation of the genome of the bacteriophage lambda. Q alters RNA polymerase and allows transcription of genes coding for heads and tails production. Original Goals
The goal of this subproject is to determine the approximate concentration of Q necessary to effectively switch from a lysogenic to a lytic pathway. Q amber mutants, which are deficient in the production of Q, will be used, and the production of Q in the host cell will be controlled by varying the concentration of inducer ATC. The fraction of colonies that lyse will be measured, and a graph of lysis vs. Q production will be obtained.
Status and Future Plans
The Q constructs, consisting of promoter pTet, spacer, Q gene, double terminator, constitutive promoter, tetR gene, and double terminator have been made and sequence verified. There are two version of the construct, one with a medium-strength constitutive promoter driving tetR, and one with a strong constitutive promoter. Titration trials with Q amber mutants have shown that expression of Q under tetR repression using the medium-strength promoter is leaky but inducible with anhydrotetracycline (aTc). Replacing the spacer with various cis-repressor sequences has shut down the leaky repression.
Future work on Q includes titering the construct featuring the strong constitutive promoter, introducing the conjugate trans activators to Q construct variants containing cis repressors, and quantitatively measuring the effects of aTc concentration on cell lysis rates.
The Q construct used to determine concentration of Q necessary for lytic pathway induction.
|