Cambridge/Gram-positive chassis project
From 2007.igem.org
(→Details) |
(→Research on ''B. subtilis'') |
||
| Line 34: | Line 34: | ||
== Research on ''B. subtilis'' == | == Research on ''B. subtilis'' == | ||
| - | Check out the papers we've trawled through [ | + | Check out the papers we've trawled through [http://www.ccbi.cam.ac.uk/iGEM2007/index.php/Detailed_background_information_-_B._subtilis here]. |
== Acquiring our Bacillus strains: the ''B. subtilis'' shopping list == | == Acquiring our Bacillus strains: the ''B. subtilis'' shopping list == | ||
Revision as of 22:42, 25 October 2007
Background
Bacillus subtilis is a Gram-positive bacterium commonly found in the soil. B. subtilis is not considered a human pathogen - it might contaminate food but reports of food poisoning are very rare. The terms Gram-positive and Gram-negative refer to a major subdivision of bacteria into two classes, based on how they react to a particular staining method, with important implications for their cell strucutre - see the [http://en.wikipedia.org/wiki/Gram_staining Wikipedia article on the Gram stain] for a more detailed explanation.
As genetic manipulation of B. subtilis can be done rather easily, it is thus widely adopted as a model organism for laboratory studies, especially of sporulation, which is a simplified example of cellular differentiation.
While the E. coli Gram-negative chassis has been very well characterized and widely used in the synthetic biology world, there is no such chassis available for Gram-positive bacteria. This year we intend to create a B. subtilis chassis and make BioBricks for it available in the [http://partsregistry.org/Main_Page MIT Standard Registry of Parts], where it will be freely available to future iGEM teams and interested researchers alike.
Motivation
There are several reasons for using Gram-positive bacteria in general, and B. subtilis in particular, for synthetic biology (as well as/instead of the usual Gram-negative E. coli strains):
- Since they do not have a second membrane, they will secrete and absorb substances more efficiently than Gram-negative bacteria.
- B. subtilis can be easily transformed by the natural competency method.
- B. subtilis is more motile than E. coli so will be more suitable for experiments on cell movement.
- Since B. subtilis be easily cultured along the lines of standard E. coli protocols, not much new equipment will be required.
- B. subtilis is a Class I contaminant ([http://www.epa.gov/oppt/biotech/pubs/fra/fra009.htm US EPA webpage]), so it can be used without potential health or environmental risks.
As well as technical advantages of using B. subtilis, there are other ones. Firstly, we will be pioneering the use of an organism other than E. coli in the iGEM competition, which includes determining the best standard growth and transformation procedures. Secondly, we will create many biobricks for B. Subtilis such as promoters, RBS, the Agr system and the different mutants of B. subtilis. Since currently the registry of parts contains only E. coli, creating a chassis for a Gram-positive bacterium will be very valuable for the future. This will notably useful for future iGEM competitions
Details
As such, we are:
- Modifying existing B. subtilis integrational and shuttle plasmids to have the required BioBrick restriction sites, so that BioBricks can be first assembled in a suitable E. coli chassis and then transformed into B. subtilis
- Investigating experimental protocols for dealing with B. subtilis
- Characterizing the strength of present E. coli promoters in B. subtilis
- Creating BioBricks of promoters which work very well in both E. coli and B. subtilis
- Contributing new integrational vectors that will allow the insertion of novel genes into the B. subtilis host
It is hoped that our work will pave the way for the use of B. subtilis amongst synthetic biologists, iGEM in paricular, and that the problems we face along the way be of reference to other groups working on B. subtilis in the future.
In addition, for the signalling project we intend to transfer our agr BioBricks into B. subtilis and later, using the FepA permeability device, into E. coli - if successful, this will allow cross-talk between two very different bacterial species.
Research on B. subtilis
Check out the papers we've trawled through [http://www.ccbi.cam.ac.uk/iGEM2007/index.php/Detailed_background_information_-_B._subtilis here].
Acquiring our Bacillus strains: the B. subtilis shopping list
Which mutants?
- Sporulation mutants are useful in preventing the killing of E. coli by B. subtilis if they're cultured together.
- Antibiotic production in the bacilli is accompanied by cessation of vegetative growth and spore formation [http://textbookofbacteriology.net/Bacillus.html], hence sporulation mutants won't produce antibiotics (hopefully)
- Protease mutants would (hopefully) increase AIP yields, no papers to justify this however.
Strains ordered
All these strains are in the Bacillus stock centre ([http://www.bgsc.org BGSC]). The following numbers are used by BGSC:
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1a1 1a1] (B. subtilis strain 168)
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1s15 1s15] (spo0A322 - whatever the "322" means)
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1a751 1a751] (aprE - a protease)
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=ECE10 ECE10] (contains the pBS42 shuttle vector)
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=ECE62 ECE62] (contains pRB373)
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=ECE63 ECE63] (contains pRB374)
All of these can be cultured on LB; ECE10 requires LB + chloramphenicol while ECE62 and ECE63 requires LB + ampicillin to select for cells with the plasmids.
Strains received
Our FedEx-ed shipment arrived 2nd August, and on top the strains ordered by us, the director kindly attached the following strains, which he personally recommends for our use:
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=ECE168 ECE168] (contains pMTLBs72 shuttle vector)
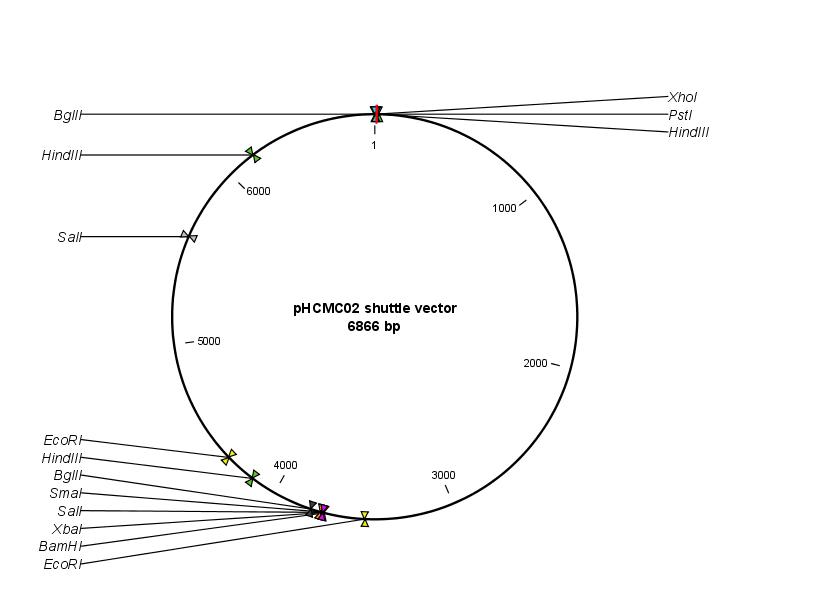
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=ECE188 ECE188] (contains pHCMC02 shuttle vector)
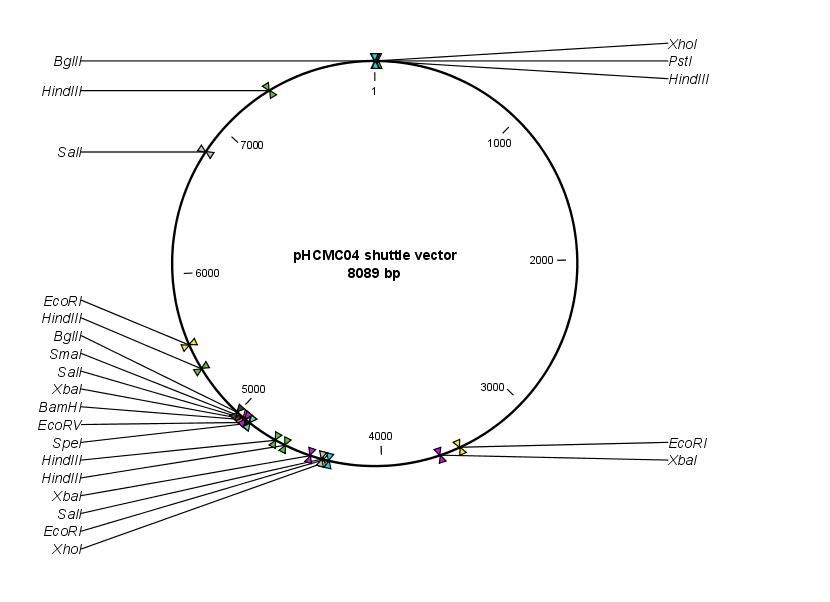
- [http://newman.uts.ohio-state.edu/BGSC/getdetail.cfm?bgscid=ECE189 ECE189] (contains pHCMC04 shuttle vector)
In addition, we were given the following plasmids and E. coli host by Jan-Willem Veening from the Errington lab at the University of Newcastle:
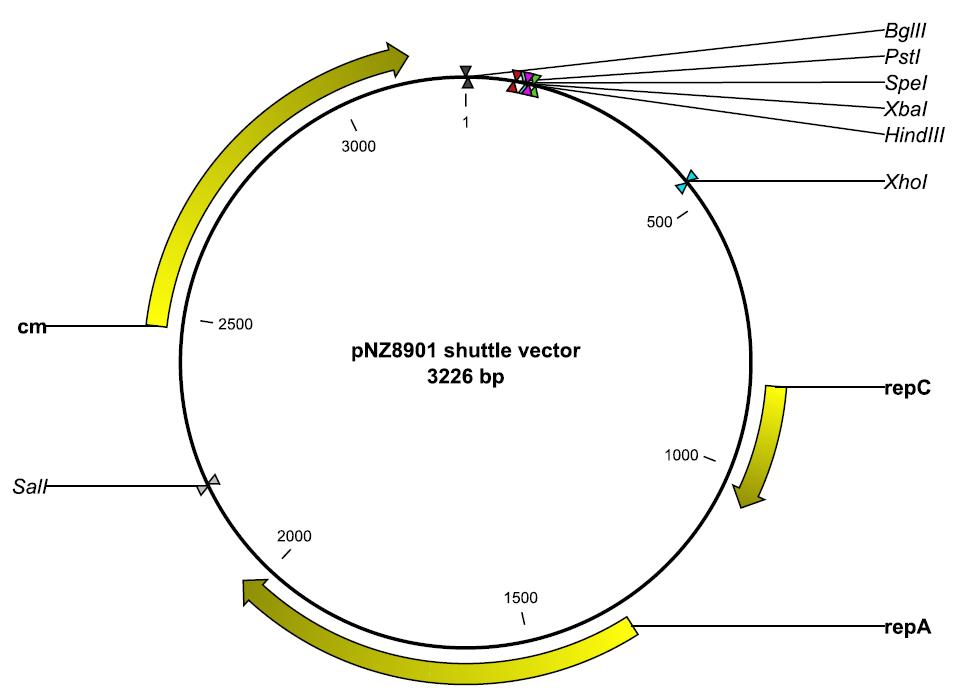
- pNZ8901
- pPL82
- E. coli strain MC1061
Plasmids not tested on
These plasmids were not experimented on as they had either EcoRI, XbaI, SpeI or PstI cutting sites present outside of the multiple cloning site.
|
|
Also, pMTLBs72 was ruled out because in the primary paper ([http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WPF-47RRT96-2&_user=1495569&_coverDate=01%2F31%2F2003&_fmt=full&_orig=search&_cdi=6989&view=c&_acct=C000053194&_version=1&_urlVersion=0&_userid=1495569&md5=66e7d231d9bdaf940b3beeb2d0aa8ddd&ref=full]) that described its construction, they had a link to BGSC for the plasmid map, but it's not on BGSC either.
Testing of vectors
Summary table of vectors tested
| Plasmid | Criterion | ||
|---|---|---|---|
| Sequence known? | Growth in E. cloni | Growth in B. subtilis | |
| pRB373 | No (only plasmid map available [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=1452028&dopt=Citation here]. Paper yet to be digitized) | Yes | No |
| pBS42 | Unsure, discussed in next section (GenBank format, pdf) | Yes | Yes (but not when it's modified) |
| pNZ8901 | Yes (GenBank format, pdf) | Yes (2 days) | Yes (Overnight) |
| pPL82 | Yes (Similar to pDR67, pdf) | Yes (2 days) | Yes (Overnight) |
Reasons for stopping work on..
pRB373 and pRB374
pRB373 and pRB374 were shipped pre-transformed into E. coli cells. Transformation into Bacillus strain 1A1 did not succeed with either plasmid and this corroborates with Dr Zeigler (BGSC director)'s comments that previous users have reported difficulty in getting them to replicate in Bacillus.
pBS42
- Conflicting sequence data and restriction enzyme maps - the singly-digested plasmid produced a band of length 5.6kb, similar to [http://seq.yeastgenome.org/vectordb/vector_descrip/COMPLETE/PBS42.SEQ.html this sequence], but restriction enzyme sites resembled [http://seq.yeastgenome.org/vectordb/vector_descrip/COMPLETE/PBS42_SUBMITTED.SEQ.html this sequence] instead, but that is 4.5kb only.
- The region that was removed from the plasmid so that we could insert BioBricks in was originally from pUB110 (a Bacillus plasmid). This section might have contained the origin of replication for Bacillus, thus the lack of colonies when the modified pBS42 was transformed into Bacillus.
Description of experimental procedures
We set out trying to determine which promoters were constitutively active in both E. coli and B. subtilis, as our final agr signalling system constructs require the constitutive expression of some genes in B. subtilis.
Introducing BioBricks into vectors
Promoters tested:
- [http://partsregistry.org/Part:BBa_I13604 BBa_I13604]: Tet operator ([[http://partsregistry.org/Part:BBa_R0040 BBa_R0040]) + YFP(LVA-) reporter, followed by a Lac operator ([http://partsregistry.org/Part:BBa_R0011 BBa_R0011]) + CFP(LVA-) reporter
- [http://partsregistry.org/Part:BBa_I13973 BBa_I13973]: cI promoter ([http://partsregistry.org/Part:BBa_R0051 BBa_R0051]) + YFP-aav reporter device
- [http://partsregistry.org/Part:BBa_J04450 BBa_J04450]: RFP Coding Device switched on by IPTG, contains LacI promoter ([http://partsregistry.org/Part:BBa_R0010 BBa_R0010])
- [http://partsregistry.org/Part:BBa_I746105 BBa_I746105]: GFP with agr P2 promoter ([http://partsregistry.org/Part:BBa_I746104 BBa_I746104])
- Note: the agr P2 promoter is derived from S. aureus, but we were unsure whether it would work in B. subtilis or E. coli as it was never previously reported in literature.
In order to test if promoters in the BioBrick registry functioned in Bacillus, this was the protocol we adopted for the testing:
| Inserts | Vectors |
|---|---|
|
|
| and then.. | |
| |
Checking function of promoters in B. subtilis
For pBS42:
- Fluorescent E. cloni colonies were obtained for BBa_I13604 and BBa_J04450. After miniprepping and transformation into Bacillus, no transformants were obtained. A possible reason could be that part of the Bacillus origin was removed during the process of constructing the vector+insert as according to the preliminary sequence, the region removed was from the pUB110 plasmid (1-2600).
For pNZ8901:
- Fluorescent E. cloni colonies were obtained for BBa_I13604, BBa_J04450 and BBa_I746105. After miniprepping and transformation into Bacillus, none of the colonies showed fluorescence. In the case of BBa_J04450, after miniprepping the Bacillus colonies and running a double digest to separate insert from vector, some colonies were found to contain 2 bands of the correct size, suggesting that the greater stringency behind use of promoters in Bacillus may be the cause. No DNA was recovered from the miniprep of pNZ + BBa_I13604.
For pPL82:
- Fluorescent E.coli colonies were obtained for BBa_I13604, BBa_J04450 and BBa_I746105. After miniprepping and transformation into B.subtilis, none of the colonies showed fluorescence. However, this doesn't provide a reliable test for promoter efficiency in B.subtilis as pPL82 works by integrating into the B.subtilis chromosome, which effectively renders it single copy. Also, unlike the episomal plasmids, successful transformation of pPL82 can't be checked by miniprepping as the genes on the plasmid integrate into the bacterial chromosome.
Modification of vectors into standard BioBrick chasses #1: pNZ8901
Preparation step
- Two primers were ordered
- NsiI + BamHI + [http://partsregistry.org/Part:BBa_G00100 VF2] (GTC ATG CAT GGA TCC TGC CAC CTG ACG TCT AAG AA)
- HindIII + SalI + [http://partsregistry.org/Part:BBa_G00101 VR] (GTC AAG CTT GTC GAC ATT ACC GCC TTT GAG TGA GC)
- pNZ8901 transformed into E. cloni, plasmids miniprepped subsequently.
- [http://partsregistry.org/Part:BBa_E0840 E0840] transformed into E. cloni, plasmids miniprepped subsequently.
Basis for experiment
- When PCR is carried out using E0840 as template with the two primers detailed above, a linear piece of DNA of the following sequence would be produced:
NsiI BamHI EcoRI XbaI SpeI PstI SalI HindIII ---|------|---------|------|---[GFP reporter construct]---|------|--------|------|---
- From literature research, it was concluded that the region between BglII and XhoI does not contain important genes pertaining to plasmid replication or copy number control.
- There is a terminator sequence near the 300bp mark, which is intended to stop expression of genes cloned into the MCS
- The restriction endonucleases BamHI and BglII produces the same 4bp overhangs (sticky ends) after digestion; SalI and XhoI produces complementary overhangs as well
- The ligation of a BamHI and BglII cut site would produce a scar (conceptually similar to the scar produced when XbaI ends and SpeI ends ligate together during standard BioBrick assembly); so would SalI-XhoI ligations.
- This effectively reduces the number of available cutting sites in the plasmid - standard chasses tend to have very few cutting sites beyond the ones required for standard assembly.
- As backup (and to save cost), in case the region bounded by BglII and XhoI was important, we decided it would be good to choose two other cutting sites which were nearer to each other.
- NsiI-PstI ligations would produce a scar, but as HindIII do not have other complementary enzymes, it was decided that HindIII should be kept intact in the final plasmid.
Experiments carried out, round I
- PCR was carried out according to the outline above
- PCR mixture were double digested with BamHI and SalI
- Double digested mixture cleaned (to remove the smaller bits chopped off), DNA run on gel to analyze concentrations
- Worked pretty well, size of bands were correct
- Plasmid double digested with BglII and XhoI
- Gel extraction carried out to purify cut vector
- Ligation of insert with cut vector left overnight
- Transform ligation mixture into E. cloni, grow on Chl plates, culture, miniprep.
- Digest the miniprepped DNA with EcoRI and other restriction enzymes as preliminary check for the correct plasmid.
Digests with XbaI, SpeI and PstI revealed that the plasmid was of the correct size (slightly over 4kb), but EcoRI digests produced 2 fragments of 2kb each. From not having a single EcoRI cutting site, the plasmid now has two. All of the colonies picked and miniprepped show the same band patterns. Might it be because PCR wasn't that accurate?
Experiments carried out, round II
The backup plan was put into action. PstI and HindIII was used for the plasmid; NsiI and HindIII for the PCR-ed DNA. Other steps remain the same as described above.
Colony PCR (with plain VR and VF2 primers) indicated that the colonies had the inserts, but after culturing and miniprepping, the DNA vanished into thin air. That, mystifyingly, happened to all of the cultured colonies.
Experiments carried out, round III
The steps used in round I and II were carried out again. This time round, colony PCR failed to show any transformants having the desired vector, even when 30 colonies in total were picked.
Conclusion
Although we have yet to modify pNZ8901 to conform to having the standard cutting sites similar to standard BioBrick chasses available through the Registry, we have devised a method to modify pNZ8901 that should work if we were a bit more lucky and had more time. This method would also be applicable to other plasmids that other groups might want to contribute to the Registry.
Modification of vectors into standard BioBrick chasses #2: pPL82
Knocking out of PstI site outside of MCS
The original pPL82 contains a PstI site outside the MCS within the Amp resistance gene. Site-directed mutagenesis was carried out to knock out this PstI site:
- pPL82 was digested with PstI
- Restriction enzyme was inactivated by QIAGEN restriction digest clean up kit
- PstI site knock out was carried out with T4 DNA polymerase by incubating the reaction mixture together with T4 DNA Polymerase first in the absence of dNTP for 10 minutes followed by incubation with dNTP addition for 20 minutes
- Reaction mixture was cleaned up again following step 2 above
- Blunt end ligation was carried out for 1 hour with T4 DNA ligase at room temperature
- Transformation of E. coli was carried out with electroporation, with chloramphenicol selection
- The successful transformants were characterized for Amp resistance. Successful PstI site knock outs are expected to be sensitive to Amp, as 4 base pairs would have been lost during the mutagenesis process due to the action of the exonuclease activity of the T4 DNA polymerase. This would give a frame shift in the translation of the Amp Resistance gene.
Introducing a SpeI and PstI site in the MCS
The PstI knock out pPL82 vector is then double digested with EcoRI and Xbal, and then the vector backbone is gel extracted using QIAGEN GEl Extraction kit (9kb backbone + 300bp fragment).
To modify pPL82's MCS into the standard prefix and suffix configuration, 2 single stranded DNA oligos are ordered that corresponding to the forward and reverse strand of --X--S-P-. In order to ligate this insert into the double digested pPL82 backbone, sticky end corresponding to EcoRI and SpeI site are added to each side of the oligo respectively. Ligation of the sticky EcoRI site on one side of the oligo with the digested pPL82 backbone would restore the EcoRI site; while ligation between the sticky SpeI site with the digested Xbal site on the vector would produce a scar that destroys both sites.
Hence, the modified vector would contain the standard -E-X--S-P- that allows for standard assembly and verification using colony PCR with the corresponding prefix and suffix primers.
forward single stranded oligo: 5' AATTCGCGGCCGCTTCTAGAGTACTAGTAGCGGCCGCTGCAGA 3'
|||||||||||||||||||||||||||||||||||||||
reverse single stranded oligo: 3'GCGCCGGCGAAGATCTCATGATCATCGCCGGCGACGTCTGATC 5'
The 2 single stranded oligo are reanealed by mixing 2ul of each from their 10uM stock together with 16ul of SDW in a eppendorf tube on boiling water. The mixture is allowed to cool down to room temperature slowly before added to the ligation mixture with the prepared pPL82 E/X gel extraced vector backbone. Standard ligation and transformation follows.
Conclusions
Achievements
We can say for sure that we have successfully introduced J04450, containing R0010 (the lacI promoter), into B. subtilis, and it did not work. The problem lies more with the promoter than the fluorescent proteins, as other fluorescent proteins (GFP, CFP, YFP) have been shown to work in B. subtilis [http://aem.asm.org/cgi/content/full/68/5/2624?view=long&pmid=11976148].
For the other promoters, we have attempted to introduce it into B. subtilis but had never observed expression of fluorescent proteins, and have yet to be able to confirm that the introduced plasmid (which conferred antibiotic resistance) could be stably inherited.
We have created a new promoter, Pspac-hy, which is recognised by both the E. coli and B. subtilis transcription machinery according to our literature searches. We confirmed that it was able to drive GFP expression in a standard BioBrick plasmid (pSB1A2) in E. coli but have yet to be introduced into B. subtilis via the shuttle vector for checking.
Problems faced
We would like to highlight the possible difficulties one might run into when working with B. subtilis as a chassis:
Slow growth of E. coli cells containing pNZ8901 and pPL82
One unforseen, and proved to be highly detrimental to our efficiency was the slow growth of pNZ8901 and pPL82 in E. cloni, our preferred E. coli chassis. It takes upwards of 36 hours for a liquid culture broth of E. cloni containing either plasmid to turn cloudy; similarly, these E. cloni cells take approximately the same time to produce visible colonies on an agar plate. This is because the origin of replication is optimized for B. subtilis and less for E. coli.
However, when these plasmids were transformed into B. subtilis, colonies and liquid culture broths grew to satisfactory levels overnight.
Difficulty in miniprepping DNA from B. subtilis cells
After successfully transforming B. subtilis cells with pNZ8901 containing the promoter of interest + GFP construct, despite not observing fluorescence from cells that exhibit resistance to chloramphenicol (encoded by the plasmid), we still wanted to confirm the presence of the construct in the cells.
It took us many tries before we were able to definitively confirm the presence of the construct in B. subtilis - often the miniprepped and digested DNA, when run on an agarose gel, indicated the presence of a DNA piece larger than that of the inserted plasmid, or produced multiple bands that were not predicted by the sequence of the plasmid.
Plasmids that play hide-and-seek in E. cloni cells
Nearer to the end of our project, we had colonies that, mysteriously, did not produce DNA after miniprepping. These colonies were previously checked with colony PCR in order to confirm the existence of the construct of interest, and PCR results show that those cells had the plasmid with the construct of interest.
Regarding the reasons behind the failure for the plasmids to be miniprepped out, and why it happened not just once or twice, remains a mystery. Might it be possible that the plasmid DNA integrated into the E. coli chromosome?
Unoptimized transformation protocols for B. subtilis
Initially, we gleaned B. subtilis transformation protocols from the internet, as well as seeking help on transformation protocols from a Bacillus lab in Newcastle, but both protocols did not give satisfactory levels of transformation in our B. subtilis plates.
We have thus carried out an assay of different conditions during the transformation of B. subtilis, and one of our important findings was that the addition of EGTA led to a remarkable reduction in transformation efficiency. This conflicts with the currently-held view that the presence of chelators in the transformation step increased efficiency of transformation ([http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=16561900&dopt=Citation]).
For more information, check out the section on the assay of transformation protocols below.
Potential for further work
Modification of E. coli ori in vectors
As the introduction of pNZ8901 and pPL82 hampered the growth of the E.coli host and hence the efficiency of our experiments, it would be a great idea to modify the ori to one which is optimized for E. coli as well, allowing for quick overnight growth.
Identifying other suitable shuttle vectors
There are many other shuttle plasmids available in the BGSC and Bacillus labs worldwide, and it is very possible that a better episomal or integrational plasmid would emerge after more work is carried out on this area. Even so, the methods we used are still very relevant when dealing with these new plasmids - testing of promoters and modification to BioBrick standards can still be done the same way using these plasmids.
Identifying a B. subtilis strain more amenable to genetic manipulation
Our experience in introducing episomal plasmids into B. subtilis strain 168 (trpC2-) seems to indicate there is a non-negligible rate of integration of the plasmids into the bacterial chromosome, which is not surprising as Bacillus in the wild are naturally competent - they take in linear DNA from their surroundings and integrate it into their chromosomes.
The non-negligible rate of integration may help to explain the occasional absence of DNA after miniprepping, or even having multiple bands of unexpected sizes on the gel when the digested plasmid fragments are run on it.
Also, protease mutants of Bacillus strain 168 are a possible alternative host should degradation of proteins secreted into the medium be a problem.
Expanding to other Gram-positive chasses
The work done on B. subtilis can be extended to other Gram-positive bacteria such as other Lactobacilli and Streptococci genus, which might turn out to be a better understood chassis than B. subtilis in the years to come.
Adaptation for industrial purposes
Using the integrational vector, genes encoding proteins of pharmaceutical/industrial importance(such as insulin) can be introduced stably into a Gram-postive chassis and the product secreted directly into the medium, which would prove to be more efficient than the conventionally-used Gram-negative chassis.
Acknowledgements
We would like to thank:
- [http://www.bgsc.org BGSC] for providing us the B. subtilis strains and plasmids used during the project
- The Errington lab in Newcastle University, especially J-W. Veening who sent us pNZ8901 and pPL82, and also gave us valuable insights throughout the project
References
Key papers used:
- Bruckner R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene. 1992 Dec 1;122(1):187-92. [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=1452028&dopt=Citation]
- Anagnostopoulos C, Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741-6. [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=16561900&dopt=Citation]
- Yansura DG, Henner DJ. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Jan;81(2):439-43. [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=retrieve&db=pubmed&list_uids=6420789&dopt=Citation]
- Bongers RS, Veening JW, Van Wieringen M, Kuipers OP, Kleerebezem M. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl Environ Microbiol. 2005 Dec;71(12):8818-24. [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1317459]
For a more detailed view of our research, please visit Detailed background information - B. subtilis
Postscript: The great Bacillus competency/transformation assay
Introduction
We were mystified as to why the transformation efficiency of our plates were not as high as expected, casting doubt on the protocols we have been faithfully following on our own wiki, which is obtained from an online forum.
We have received a lot of help from a Bacillus lab in Newcastle - they sent us the protocol they use in making B. subtilis competent, as well as their transformation protocol. Both protocols were largely similar, hence the resulting protocol used in the assay is a merged version of both protocols.
For experimental details, please refer to the article on the great Bacillus competency/transformation assay.
Conclusions
- Addition of Trp resulted, in most cases, a slight increase in the number of colonies on the plate
- Addition of EGTA led to a drastic decrease in the number of colonies obtained
- 6x and 9x are considered to be optimal concentrations for dilution; as can be seen, 3x and 12x has less colonies, especially 3x. This shows that B. subtilis is competent only at a certain stage of its growth (log phase?)
- The variability in the frozen cells assay is attributed to the uneven mixing of the DNA into the 1ml culture, and the subsequent uneven aliquoting of 504ul of the mixture into the second Eppendorf tube.
- Comparing 10%(v/v) and 30%(v/v) plates, the difference is not significant, but there seems to be more colonies on the 10%(v/v) plate
- Comparing the thawed cells to the fresh cells, it seems as if the difference is not significant as well - blanket colonies are still possible
The 4 Bacillus commandments
- Thou shalt dilute cells to 9x concentration after the initial culturing step
- Take OD450 readings at the start and end of the culture phase to check that the cells are in the correct phase of growth for transformation.
- Thou shalt add Trp, but if thou forgets, it is of not of large consequence
- Thou shalt never use EGTA with SpII (the starvation media)
- If needed, thou shalt freeze cells using glycerol to either 10%(v/v) or 30%(v/v)