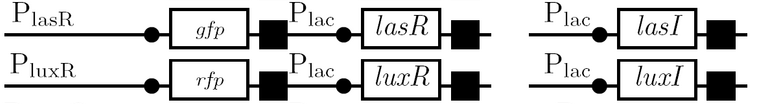Construction and Testing
From 2007.igem.org
(Difference between revisions)
(→Construction Tree) |
|||
| Line 3: | Line 3: | ||
[[Image:UW_Construction_Chart.jpg|thumb|500px|left|Construction Chart]] | [[Image:UW_Construction_Chart.jpg|thumb|500px|left|Construction Chart]] | ||
Diagram details: | Diagram details: | ||
| - | *Vector resistances are shown, with shades of blue representing ampicillin and shades of red representing kanamycin | + | *Vector resistances are shown, with shades of blue representing ampicillin and shades of red representing kanamycin. |
| - | * | + | *Parts and developing constructs all have abbreviated names for ease of labeling. To make the subtle distinction between a coding sequence and a coding sequence with a ribosome binding site, we use a ' on the computer and an underline when writing. A double ' or double underline indicates a coding sequence with a ribosome binding site and a transcriptional terminator. |
*Expected fragment sizes are shown at every node so gel results can be analyzed quickly. | *Expected fragment sizes are shown at every node so gel results can be analyzed quickly. | ||
| - | *Arrows show | + | *Arrows show the direction of DNA cloning and specify which restriction enzymes to use for cutting. Hollow arrows are present when two parts are ligated from vectors of the same resistance, requiring additional methods such as the use of alkaline phosphatase, gel extraction, or a three-way ligation into a vector of difference resistance. |
| - | *The numbers 1-2-3-3.5-4 show the major | + | *The numbers 1-2-3-3.5-4 show the major protocols that need to be carried at each step in the assembly cycle. |
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
Revision as of 13:10, 26 October 2007
Contents |
Construction Tree
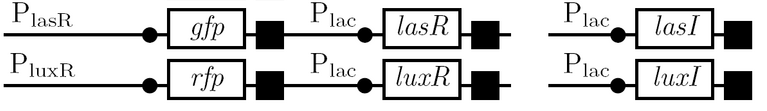
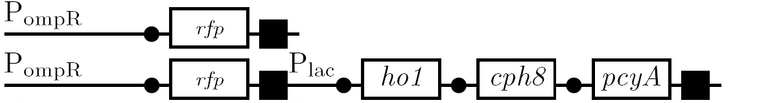
The diagram below outlines our parallel construction plan, from individual registry parts to the final half-adder. Because many people were working on the project at once, it is designed to keep everyone organised.
Diagram details:
- Vector resistances are shown, with shades of blue representing ampicillin and shades of red representing kanamycin.
- Parts and developing constructs all have abbreviated names for ease of labeling. To make the subtle distinction between a coding sequence and a coding sequence with a ribosome binding site, we use a ' on the computer and an underline when writing. A double ' or double underline indicates a coding sequence with a ribosome binding site and a transcriptional terminator.
- Expected fragment sizes are shown at every node so gel results can be analyzed quickly.
- Arrows show the direction of DNA cloning and specify which restriction enzymes to use for cutting. Hollow arrows are present when two parts are ligated from vectors of the same resistance, requiring additional methods such as the use of alkaline phosphatase, gel extraction, or a three-way ligation into a vector of difference resistance.
- The numbers 1-2-3-3.5-4 show the major protocols that need to be carried at each step in the assembly cycle.
Testing Constructs
Making use of flurorescent proteins(GFP and RFP) as reporter genes - use of indicuble promoters to...
Comparing basal and induced expression levels for the quorum sensing promoters
Testing functionality of the various promoters
Home