Davidson Missouri W
From 2007.igem.org
(→'''Students''') |
Macampbell (Talk | contribs) (→Our Successful Project) |
||
| (220 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | ===Davidson | + | <center>[[Davidson Missouri W| <span style="color:black">Home</span>]] | [[Davidson Missouri W/Background Information| <span style="color:red">Background Information</span>]] | [[Davidson Missouri W/Solving the HPP in vivo| <span style="color:red">Current Project: Solving the Hamiltonian Path Problem ''in vivo''</span>]] | [[Davidson Missouri W/Mathematical Modeling| <span style="color:red">Mathematical Modeling</span>]] | [[Davidson Missouri W/Gene splitting| <span style="color:red">Gene Splitting</span>]] | [[Davidson Missouri W/Results| <span style="color:red">Results</span>]] | [[Davidson Missouri W/Traveling Salesperson Problem| <span style="color:red">Traveling Salesperson Problem</span> ]] | [[Davidson Missouri W/Software|<span style="color:red">Software</span>]] | [[Davidson Missouri W/Resources and Citations|<span style="color:red">Resources and Citations</span>]]</center> |
| - | [[ | + | |
| + | <hr> | ||
<br> | <br> | ||
| + | [[Image:dmw_logo2.png|center]] | ||
| - | + | [[Image:Computer.png|center]] | |
| - | + | ||
| - | + | <center> | |
| + | =The Team= | ||
| + | </center> | ||
| + | {| border="1" cellpadding="5" cellspacing="0" align="center" width="100%" | ||
| + | |- | ||
| + | ! style="color: white; background-color: black;"| The Team | ||
| + | ! style="color: white; background-color: black;" | The Faculty | ||
| + | ! style="color: white; background-color: black;" | School Logos | ||
| + | ! style="color: white; background-color: black;" | Group Photo | ||
| + | |- | ||
| - | + | |style="color: black; background-color: red;" align="center"| '''Davidson''' | |
| + | <b> | ||
| + | [[Davidson Missouri W/Oyinade Adefuye|<span style="color:black">Oyinade Adefuye</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Will DeLoache|<span style="color:black">Will DeLoache</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Jim Dickson|<span style="color:black">Jim Dickson</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Andrew Martens|<span style="color:black">Andrew Martens</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Amber Shoecraft|<span style="color:black">Amber Shoecraft</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Mike Waters|<span style="color:black">Mike Waters</span>]] | ||
| + | </b> | ||
| - | + | |style="color: black; background-color: red;" align="center"| | |
| + | <b> | ||
| + | [[Davidson Missouri W/A. Malcolm Campbell|<span style="color:black">A. Malcolm Campbell</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Karmella Haynes|<span style="color:black">Karmella Haynes</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Laurie Heyer|<span style="color:black">Laurie Heyer</span>]] | ||
| + | </b> | ||
| - | + | |style="color: black; background-color: white;" align="center"| | |
| + | [[Image:DavidsonLogo.gif]] | ||
| - | + | |style="color: black; background-color: white;" align="center"| | |
| + | [[Image:Team1.jpg|thumb|center|300px]] | ||
| - | + | |- | |
| - | + | |style="color: black; background-color: gold;" align="center"|'''Missouri Western''' | |
| + | <b> | ||
| + | [[Davidson Missouri W/Jordan Baumgardner|<span style="color:black;">Jordan Baumgardner</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Tom Crowley|<span style="color:black;">Tom Crowley</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Lane H. Heard|<span style="color:black;">Lane H. Heard</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Nickolaus Morton|<span style="color:black;">Nickolaus Morton</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Michelle Ritter|<span style="color:black;">Michelle Ritter</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Jessica Treece|<span style="color:black;">Jessica Treece</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Matthew Unzicker|<span style="color:black;">Matthew Unzicker</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Amanda Valencia|<span style="color:black;">Amanda Valencia</span>]] | ||
| + | </b> | ||
| - | + | |style="color: black; background-color: gold;" align="center"| | |
| + | <b> | ||
| + | [[Davidson Missouri W/Todd Eckdahl|<span style="color:black;">Todd Eckdahl</span>]] | ||
| + | <br> | ||
| + | [[Davidson Missouri W/Jeff Poet|<span style="color:black;">Jeff Poet</span>]] | ||
| + | </b> | ||
| - | + | |style="color: black; background-color: white;" align="center"|[[Image:MWLogo.gif]] | |
| - | + | |style="color: black; background-color: white;" align="center"|[[Image:MWSUteam.jpeg|thumb|center|300px]] | |
| + | |- | ||
| - | + | |} | |
| + | <br> | ||
| + | <center> | ||
| - | + | =Our Successful Project= | |
| + | </center> | ||
| - | ''' | + | {| border="1" cellpadding="5" cellspacing="0" align="center" width="90%" |
| + | |- | ||
| + | ! style="color: black; background-color: red;" width="20%"| <font size="+1">In Depth</font> | ||
| + | ! colspan="3" style="color: black; background-color: red;" width="60%"| <font size="+1">Overview</font> | ||
| + | |- | ||
| + | |style="color: black; background-color: black;" align="center"| | ||
| + | [[Davidson Missouri W/Background Information|<span style="color:red">Background Information</span>]] | ||
| + | <br><br><br> | ||
| + | [[Davidson Missouri W/Solving the HPP in vivo|<span style="color:red">Current Project: Solving the Hamiltonian Path Problem ''in vivo''</span>]] | ||
| + | <br><br><br> | ||
| + | [[Davidson Missouri W/Mathematical Modeling|<span style="color:red">Mathematical Modeling</span>]] | ||
| + | <br><br><br> | ||
| + | [[Davidson Missouri W/Gene splitting|<span style="color:red">Gene Splitting</span>]] | ||
| + | <br><br><br> | ||
| + | [[Davidson Missouri W/Results|<span style="color:red">Results</span>]] | ||
| + | <br><br><br> | ||
| + | [[Davidson Missouri W/Traveling Salesperson Problem|<span style="color:red">Traveling Salesperson Problem</span>]] | ||
| + | <br><br><br> | ||
| + | [[Davidson Missouri W/Software|<span style="color:red">Software</span>]] | ||
| + | <br><br><br> | ||
| + | [[Davidson Missouri W/Resources and Citations|<span style="color:red">Resources and Citations</span>]] | ||
| + | <br><br><Br> | ||
| + | |Hamiltonian Path Problem | ||
| + | As a part of iGEM2006, a combined team from Davidson College and Missouri Western State University reconstituted a hin/''hix'' DNA recombination mechanism which exists in nature in ''Salmonella'' as standard biobricks for use in ''E. coli''. The purpose of the 2006 combined team was to provide a proof of concept for a bacterial computer in using this mechanism to solve a variation of The Pancake Problem from Computer Science. This task utilized both biology and mathematics students and faculty from the two institutions. | ||
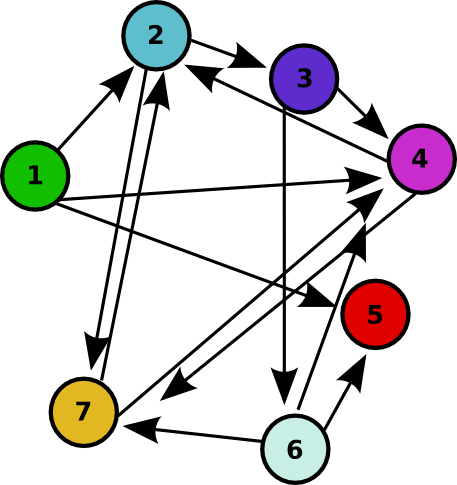
| - | + | For 2007, we successfully continued our collaboration and our efforts to manipulate ''E. coli'' into mathematics problem solvers as we refine our efforts with the hin/''hix'' mechanism to explore another mathematics problem, the Hamiltonian Path Problem. This problem was the subject of a groundbreaking paper by Adleman in 1994 (see [[Davidson_Missouri_W/Resources_and_Citations | citations]]) where a unique Hamiltonian path was found ''in vitro'' for a particular directed graph on seven nodes. We were able to use bacterial computers to solve the Hamiltonian path problem ''in vivo''. ([[Davidson Missouri W/Background Information#Why Use Bacteria?|Why use a bacterial computer?]]) | |
| - | + | <br> | |
| + | |||
| + | [[Image:Adelman.png|thumb|300px|center|The Adleman graph.]] | ||
| - | [ | + | <center> For the graph used in Adleman's paper (shown above), the Hamiltonian Path Problem would ask: can you find a path along the directed edges that travels from node 1 (green) to node 5 (red) and visits each node on the graph exactly once? <br> |
| + | [https://static.igem.org/mediawiki/2007/6/6f/Adelmansolution.png Click here] for the solution. | ||
| + | </center> | ||
| + | |} | ||
| + | <br> | ||
| + | <center> '''A Human Representation of the Adleman Graph. (mouse over to see the full effect)''' | ||
| - | + | <html> | |
| + | <head> | ||
| + | <link rel="stylesheet" href="https://2007.igem.org/wiki/index.php?title=User:Wideloache/igem2007.css&action=raw&ctype=text/css" type="text/css" /> | ||
| + | </style> | ||
| + | </head> | ||
| + | <body> | ||
| - | + | <div class="rollover"> | |
| + | <center> | ||
| + | <a href="#"></a> | ||
| + | </center> | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
| - | + | <br> | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <br> | |
| - | + | <hr> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <Previous Section | [[Davidson Missouri W/Background Information | Next Section>]] | |
| - | [ | + | </center> |
Latest revision as of 04:32, 27 October 2007
The Team
| The Team | The Faculty | School Logos | Group Photo |
|---|---|---|---|
| Davidson
Oyinade Adefuye
| |||
| Missouri Western
Jordan Baumgardner
| 
|
Our Successful Project
| In Depth | Overview | ||
|---|---|---|---|
|
Background Information
| Hamiltonian Path Problem
As a part of iGEM2006, a combined team from Davidson College and Missouri Western State University reconstituted a hin/hix DNA recombination mechanism which exists in nature in Salmonella as standard biobricks for use in E. coli. The purpose of the 2006 combined team was to provide a proof of concept for a bacterial computer in using this mechanism to solve a variation of The Pancake Problem from Computer Science. This task utilized both biology and mathematics students and faculty from the two institutions. For 2007, we successfully continued our collaboration and our efforts to manipulate E. coli into mathematics problem solvers as we refine our efforts with the hin/hix mechanism to explore another mathematics problem, the Hamiltonian Path Problem. This problem was the subject of a groundbreaking paper by Adleman in 1994 (see citations) where a unique Hamiltonian path was found in vitro for a particular directed graph on seven nodes. We were able to use bacterial computers to solve the Hamiltonian path problem in vivo. (Why use a bacterial computer?)
Click here for the solution. | ||
<Previous Section | Next Section>