Davidson Team's First HPP Construct
From 2007.igem.org
Wideloache (Talk | contribs) |
Wideloache (Talk | contribs) |
||
| Line 1: | Line 1: | ||
[[Image:3N graph.jpg|200px]] | [[Image:3N graph.jpg|200px]] | ||
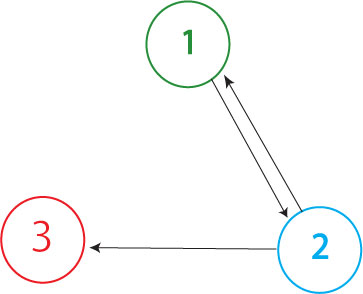
| - | The Davidson team plans to build the graph shown above in our first attempt at solving the Hamiltonian Path Problem. The Missouri Western team will build a similar graph, with an edge from node 3 to node 1 | + | The Davidson team plans to build the graph shown above in our first attempt at solving the Hamiltonian Path Problem in vivo. (The Missouri Western team will build a similar graph, with an edge from node 3 to node 1 but no edge from node 2 to node 1.) In order to determine if a Hamiltonian Path exists in the directed graph above, we propose a plasmid design similar to that shown below |
| - | + | ||
[[Image:3N solved no polymerase.jpg|800px]] | [[Image:3N solved no polymerase.jpg|800px]] | ||
| + | |||
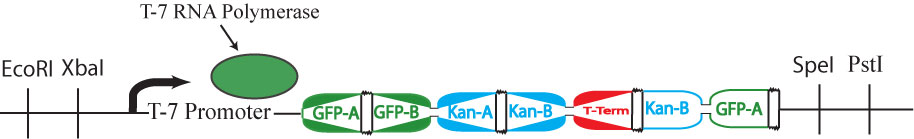
| + | This cartoon represents a solved arangement of our proposed construct. Between the two pairs of BioBrick restriction enzyme sites lies a region containing three DNA fragments, each flanked by hixC sites (represented by the black, jagged rectangles). Each node of the graph represents one of the following genes: GFP (Green Fluorescent Protein), Kanamycin Resistance and a transcription terminator. Each edge of the graph is included in the construct between two hixC sites. In the presence of Hin protein, flipping of the edges at hixC sites will produce random walks through the graph. When flipped into the correct orientation and located upstream of any transcription terminators, a given gene will be transcribed (denoted by a bold outline in the cartoon above). In the solved arrangement shown above, a Hamiltonian Path is determined to exist in the graph because all genes are expressed, and no extra edges exist in the coding region. | ||
[[Image:T7-polymerase plasmid.jpg|400px]] | [[Image:T7-polymerase plasmid.jpg|400px]] | ||
| + | |||
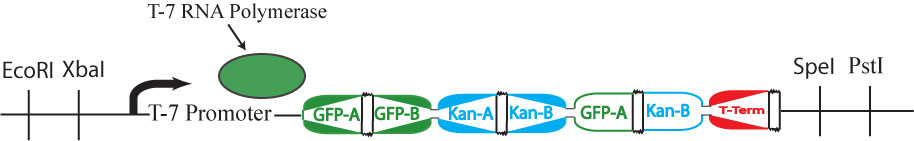
| + | It is possible for all of the genes in the graph to be expressed (or all of the nodes to be visited), but for a Hamiltonian Path to not exist in the system. A "false positive" of this type is shown above. This path (1->2->1-> through the graph is not allowed | ||
| + | |||
[[Image:3N solved polymerase.jpg|800px]] | [[Image:3N solved polymerase.jpg|800px]] | ||
[[Image:3N falsepos polymerase.jpg|800px]] | [[Image:3N falsepos polymerase.jpg|800px]] | ||
Revision as of 22:29, 28 June 2007
The Davidson team plans to build the graph shown above in our first attempt at solving the Hamiltonian Path Problem in vivo. (The Missouri Western team will build a similar graph, with an edge from node 3 to node 1 but no edge from node 2 to node 1.) In order to determine if a Hamiltonian Path exists in the directed graph above, we propose a plasmid design similar to that shown below
This cartoon represents a solved arangement of our proposed construct. Between the two pairs of BioBrick restriction enzyme sites lies a region containing three DNA fragments, each flanked by hixC sites (represented by the black, jagged rectangles). Each node of the graph represents one of the following genes: GFP (Green Fluorescent Protein), Kanamycin Resistance and a transcription terminator. Each edge of the graph is included in the construct between two hixC sites. In the presence of Hin protein, flipping of the edges at hixC sites will produce random walks through the graph. When flipped into the correct orientation and located upstream of any transcription terminators, a given gene will be transcribed (denoted by a bold outline in the cartoon above). In the solved arrangement shown above, a Hamiltonian Path is determined to exist in the graph because all genes are expressed, and no extra edges exist in the coding region.
It is possible for all of the genes in the graph to be expressed (or all of the nodes to be visited), but for a Hamiltonian Path to not exist in the system. A "false positive" of this type is shown above. This path (1->2->1-> through the graph is not allowed