Duke/Projects/efatf
From 2007.igem.org
EricJosephs (Talk | contribs) |
EricJosephs (Talk | contribs) |
||
| Line 7: | Line 7: | ||
==Parts and setup== | ==Parts and setup== | ||
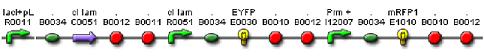
| - | We constructed a genetic circuit as an apparatus to test whether the mutant λcI repressors would bind to DNA or not under a variety of conditions. We combined [http://partsregistry.org/Part:BBa_I5010 BBa_I5010] and [http://partsregistry.org/Part:BBa_I6036 BBa_I6036] with [http://partsregistry.org/Part:BBa_I12007 BBa_I12007] and [http://partsregistry.org/Part:BBa_I13507 BBa_I13507] to yield [http://partsregistry.org/Part:BBa_S03819 BBa_S03819], shown below. | + | We constructed a genetic circuit as an apparatus to test whether the mutant λcI repressors would bind to DNA or not under a variety of conditions. We combined [http://partsregistry.org/Part:BBa_I5010 BBa_I5010] and [http://partsregistry.org/Part:BBa_I6036 BBa_I6036] with [http://partsregistry.org/Part:BBa_I12007 BBa_I12007] and [http://partsregistry.org/Part:BBa_I13507 BBa_I13507] to yield [http://partsregistry.org/Part:BBa_S03819 BBa_S03819], shown below, on an on an Amp resistant plasmid. |
[[Image:BBa_S03819.jpg]] | [[Image:BBa_S03819.jpg]] | ||
| Line 13: | Line 13: | ||
BBa_S03819 contains the λcI promotor (I12007) and repressor (R0051) sequences, nearly identical parts derived from phage λ, which give the λcI repressor protein the ability to activate or repress transcription depending on where the two sequences are placed in relation to the target protein. Thus, BBa_S03819 turns bright RED when the λcI repressor is able bound to DNA (can initiate transcription at I12007 and represses transcription at R0051) and glows YELLOW when λcI repressor is unbound (does not repress transcription at R0051 and does not activate transcription at I12007). | BBa_S03819 contains the λcI promotor (I12007) and repressor (R0051) sequences, nearly identical parts derived from phage λ, which give the λcI repressor protein the ability to activate or repress transcription depending on where the two sequences are placed in relation to the target protein. Thus, BBa_S03819 turns bright RED when the λcI repressor is able bound to DNA (can initiate transcription at I12007 and represses transcription at R0051) and glows YELLOW when λcI repressor is unbound (does not repress transcription at R0051 and does not activate transcription at I12007). | ||
| - | BBa_S03819 was then PCR'd with primers 5’-ccttggatccatAAAGAGGAGAAATACTAGATGNNNNNNNNNAAGAAACCATTAACACAAGAGCAG-3’ or 5’-ccttggatccatAAAGAGGAGAAATACTAGATGNNNNNNNNNNNNNNNCCATTAACACAAGAGCAG-3’, primers with a BamHI restriction sites with random nucleic acid N sections to yield mutants of either the first three amino acids of the flexible arm or the first five amino acids of the flexible arm, respectively, with 5’-aaggggatccgtgctcagtatcttgttatcc-3’; the PCR product was then a linear library of | + | BBa_S03819 was then PCR'd with primers 5’-ccttggatccatAAAGAGGAGAAATACTAGATGNNNNNNNNNAAGAAACCATTAACACAAGAGCAG-3’ or 5’-ccttggatccatAAAGAGGAGAAATACTAGATGNNNNNNNNNNNNNNNCCATTAACACAAGAGCAG-3’, primers with a BamHI restriction sites with random nucleic acid N sections to yield mutants of either the first three amino acids of the flexible arm or the first five amino acids of the flexible arm, respectively, with 5’-aaggggatccgtgctcagtatcttgttatcc-3’; the PCR product was then a linear library of BBa_S03819 mutated at λcI coding region. The three amino acid mutants (called in our lab 'mut3') and the five amino acid mutants ('mut5') were then digested with BamHI, ligated, transformed, and screened for DNA binding under a variety of conditions. |
==Results== | ==Results== | ||
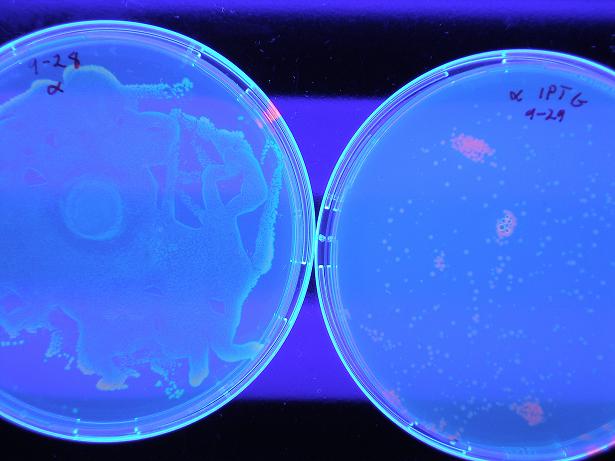
| + | Below shows BBa_S03819 both with wild-type λcI proteins unbound to DNA (not present because they are not induced with IPTG) and bound to DNA (wild type λcI proteins are induced with IPTG and present). The red fluorescence along the edges of the uninduced λcI are a result of individual bacterial colonies on the plate with naturally low LacI concentrations, so there exists λcI in the cell; this is not a concern because in screening of the mutants the λcI is always induced (grown on plates with 0.4 uM IPTG), so whether or not the present mutant λcI's bind to DNA is the only screen. Therefore, it is clear the screen works as desired, by glowing red with bound λcI and yellow (which looks green) with unbound λcI. | ||
| + | |||
| + | [[Image:BBa_S03819plate.jpg]] | ||
Revision as of 00:53, 27 October 2007
Contents |
Electric field-activated transcription factor
Background and Motivation
Experience in previous iGEM contests has shown that genetic circuits can be fickle, noisy and inhomogeneous, in contrast to electric circuits which are easily controllable but lack the breadth and variety of biological components. In order to enhance the controlability of genetic circuits, we are targeting the lifeblood of genetic circuits, the transcription factors, by attempting to guide their function with macroscopic inputs. The goal of this project is the creation of an electric-field activated transcription factor protein which will initialize transcription only in the presence of an applied, low-strength (10^3-10^5 V/m) alternating electric field.
The design aims to accomplish this goal by modifying the ‘flexible arm’ of the λcI repressor such that an applied electric field will stabilize the altered protein in an otherwise unfavorable binding with its operator site. λcI repressor This can be accomplished by screening a library of 'flexible arm' mutants, the results of which can be further screened using directed evolution techniques to yield the protein which matches the above specifications. As most proteins are fairly resistant to conformational perturbations due to electric fields , this project focuses on isolating those amino acid sequences which are particularly susceptible to electric fields.
Parts and setup
We constructed a genetic circuit as an apparatus to test whether the mutant λcI repressors would bind to DNA or not under a variety of conditions. We combined [http://partsregistry.org/Part:BBa_I5010 BBa_I5010] and [http://partsregistry.org/Part:BBa_I6036 BBa_I6036] with [http://partsregistry.org/Part:BBa_I12007 BBa_I12007] and [http://partsregistry.org/Part:BBa_I13507 BBa_I13507] to yield [http://partsregistry.org/Part:BBa_S03819 BBa_S03819], shown below, on an on an Amp resistant plasmid.
BBa_S03819 contains the λcI promotor (I12007) and repressor (R0051) sequences, nearly identical parts derived from phage λ, which give the λcI repressor protein the ability to activate or repress transcription depending on where the two sequences are placed in relation to the target protein. Thus, BBa_S03819 turns bright RED when the λcI repressor is able bound to DNA (can initiate transcription at I12007 and represses transcription at R0051) and glows YELLOW when λcI repressor is unbound (does not repress transcription at R0051 and does not activate transcription at I12007).
BBa_S03819 was then PCR'd with primers 5’-ccttggatccatAAAGAGGAGAAATACTAGATGNNNNNNNNNAAGAAACCATTAACACAAGAGCAG-3’ or 5’-ccttggatccatAAAGAGGAGAAATACTAGATGNNNNNNNNNNNNNNNCCATTAACACAAGAGCAG-3’, primers with a BamHI restriction sites with random nucleic acid N sections to yield mutants of either the first three amino acids of the flexible arm or the first five amino acids of the flexible arm, respectively, with 5’-aaggggatccgtgctcagtatcttgttatcc-3’; the PCR product was then a linear library of BBa_S03819 mutated at λcI coding region. The three amino acid mutants (called in our lab 'mut3') and the five amino acid mutants ('mut5') were then digested with BamHI, ligated, transformed, and screened for DNA binding under a variety of conditions.
Results
Below shows BBa_S03819 both with wild-type λcI proteins unbound to DNA (not present because they are not induced with IPTG) and bound to DNA (wild type λcI proteins are induced with IPTG and present). The red fluorescence along the edges of the uninduced λcI are a result of individual bacterial colonies on the plate with naturally low LacI concentrations, so there exists λcI in the cell; this is not a concern because in screening of the mutants the λcI is always induced (grown on plates with 0.4 uM IPTG), so whether or not the present mutant λcI's bind to DNA is the only screen. Therefore, it is clear the screen works as desired, by glowing red with bound λcI and yellow (which looks green) with unbound λcI.