ETHZ/Biology
From 2007.igem.org
Stefan Luzi (Talk | contribs) |
Stefan Luzi (Talk | contribs) (→Multipurpose Cell Lines =) |
||
| Line 330: | Line 330: | ||
==== Multipurpose Cell Lines ===== | ==== Multipurpose Cell Lines ===== | ||
| - | Our system can be trained to behave in a specific way by setting its inducible toggle switch to one of its two states. Let’s say this specific | + | Our system can be trained to behave in a specific way by setting its inducible toggle switch to one of its two states. Let’s say this specific states trigger specific and different events such as enzyme synthesis, transcriptional regulation, virion production or cell death. In this case one can view the bacterial cell line containing this system as a multipurpose cell line. Put another way: One has a cell line and adds a certain chemical to train it to the desired behaviour instead of constructing two independent cell lines. This means, one applies an “input engineering” instead of a “DNA engineering” approach. If one extends this idea to several inducible toggle switches being harboured in the same cell line, the number of possible phenotypes increases to 2^n, whereas n equals the number of toggle switches. E.g. if one would have 5 toggle switches inside a cell line, 32 different behaviour patterns would be possible. |
==== Intelligent Biosensors and self-adaptation ==== | ==== Intelligent Biosensors and self-adaptation ==== | ||
Revision as of 23:40, 21 October 2007

In this page, you can find an analysis of the function of our system and its relation to epigenetics, its biological design and a list of the parts that it consists of. Are you interested in constructing educatETH E.coli in your lab? Then under Lab Notes, you can find the ingredients and equipment we used, the electronic version of our lab notebook and a presentation of all the difficulties that we encountered. If you are also interested in how educatETH E.coli was modeled and simulated outside the lab, please visit the modeling and simulations web pages.
Introduction
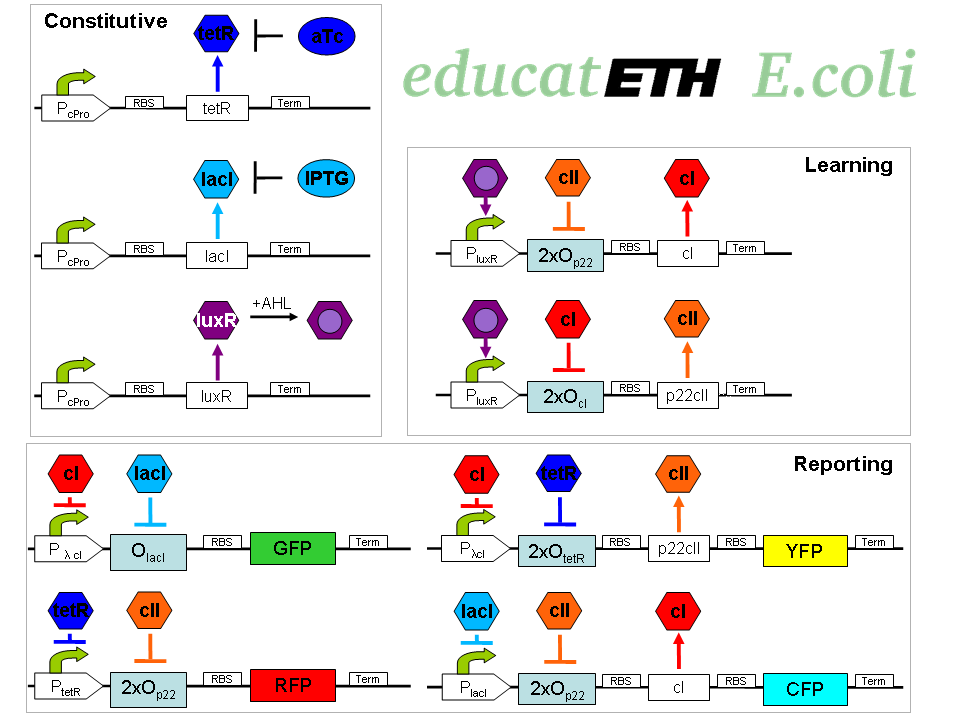
educatETH E.coli is a system which can distinguish between [http://openwetware.org/wiki/ATc anhydrotetracycline (aTc)] and [http://openwetware.org/wiki/IPTG Isopropyl-beta-D-thiogalactopyranoside (IPTG)] based on a previous training phase conducted with the same chemicals and the help of [http://partsregistry.org/Acyl-HSLs AHL]. It composes of three subsystems: the subsystem of constitutively produced proteins, the learning subsystem and the reporting subsystem. The constitutively produced proteins (LacI, TetR and LuxR) control the learning subsystem. At the core of the latter there exists a modified and extended version of the Elowitz toggle switch found in [1]. The main difference is reflected in the use of double promoters, so that the toggle switch only changes its state when both, one of the two chemicals (aTc/IPTG), and AHL are present. As AHL is only present during the training phase, the toggle maintains its state during testing, and thus can “memorize”. AHL can therefore be seen as a training- or learning substance. In the reporting subsystem, four reporters ([http://partsregistry.org/Featured_Parts:Fluorescent_proteins fluorescent proteins]) allow supervision of both the chemical the system was trained with and of if the system recognizes the chemical it is being exposed to in the testing phase as one it has been trained with or not.
The complete system
The biological design of educatETH E.coli is presented on
. In the following, we will clarify the function of all depicted components. (Are you interested in how the complex system of Fig. 1 was modelled? Then visit the Engineering Perspective!)Constitutive subsystem
The constitutively produced proteins of the system are LacI, TetR and LuxR. The LuxR part has a special function: when AHL is present, it forms a LuxR-AHL complex which acts on the learning subsystem (more on this later). At the moment, we will consider that AHL is absent and therefore LuxR cannot act on any subsystems. The TetR and LacI parts behave similarly: more specifically, the TetR protein in the absence of aTc inhibits the production of P22 cII and LacI in the absence of IPTG inhibits the production of cI. When aTc is present, however, the P22 cII production is no longer inhibited (and thus p22 cII is produced). Respectively, cI is produced when IPTG is present.
Learning subsystem
The learning subsystem is a toggle switch with two operator sites. The upper part of the toggle (cI production) has operator sites for the LuxR-AHL complex and P22 cII (which has in turn been induced by aTc). The LuxR-AHL complex induces the cI production, whereas P22 cII inhibits it. The lower part of the toggle (P22 cII production) has operator sites for the LuxR-AHL complex and cI (which has been induced by IPTG). Similarly with the upper part, the LuxR-AHL complex induces the P22 cII production and cI inhibits it. Therefore, the switch always requires the presence of the LuxR-AHL complex in order for it to operate. Its state depends on the presence of P22 cII and cI into the system, which in return was caused through the exposure of the system to aTc and IPTG.
Reporting subsystem
There are four reporters in the system. CFP (more precisely: enhanced CFP, that is eCFP) and YFP (more precisely: enhanced YFP, that is eYFP) are active during the training phase of the system and show which chemical the system is exposed to during training, whereas all four reporters (the latter and GFP and RFP) are active during the testing phase and show if the system is exposed to the same chemical as in training or not. More specifically, the YFP protein production is regulated with help of two operator sites controlled by cI and aTc. cI inhibits YFP production and aTc induces it. Therefore, YFP is produced when the system is exposed to aTc. In a similar manner, the CFP production is produced when the system is exposed to IPTG. The GFP production is regulated with help of two operator sites controlled by lacI and .
System phases
The system operation is divided into two main phases: the training phase and the testing phase. The training phase itself is also subdivided into two phases: seeing and memorizing. During seeing, the system is first exposed to one of the two chemicals it is designed to recognize (aTc and IPTG). AHL is then added and the system’s internal toggle switch reaches a steady state. During memorizing, the chemical used during seeing is removed and only AHL is retained. This maintains the toggle switch to its acquired steady state, which is reported with YFP (if aTc was seen) or CFP (if IPTG was seen). During the testing phase, the system is exposed to any of the two chemicals (aTc or IPTG), with AHL present. By comparing its toggle switch state with the effect of the newly introduced chemical, the system shows a different response if it has previously been exposed to this chemical and reports with the same XFP as in the training phase (YFP for aTc, CFP for IPTG) or if it recognizes a different chemical and reports with a different XFP (GFP for trained with aTc and recognizing IPTG, RFP for trained with IPTG and recognizing aTc).The following table presents all possible paths that may be taken by the system during all phases of operation according to the external stimuli.
| aTc | IPTG | AHL | p22cII | cI | Reporting | |
|---|---|---|---|---|---|---|
| Start | ||||||
| no input | no | no | no | no | no | no |
| Learning | ||||||
| Trained with aTc | yes | no | no | yes | no | YFP |
| Trained with IPTG | no | yes | no | no | yes | CFP |
| Memorizing | ||||||
| Trained with aTc | yes | no | yes | yes | no | YFP |
| Trained with IPTG | no | yes | yes | no | yes | CFP |
| Recognition | ||||||
| Trained with aTc Tested with aTc | yes | no | yes | yes | no | YFP |
| Trained with aTc Tested with IPTG | no | yes | yes | yes | no | GFP |
| Trained with IPTG Tested with IPTG | no | yes | yes | no | yes | CFP |
| Trained with IPTG Tested with aTc | yes | no | yes | no | yes | RFP |
Further thoughts on the system
Link to Epigenetics
Epigenetics refers to features like chromatin or DNA modifications that do not involve changes in the underlying DNA sequence and are stable over many cell divisions [1],[2]. If one has a closer look at our proposed system, one can also view it as a model-system for epigenetics: Although the DNA sequence itself stays the same, two different subpopulations of cells with different phenotypes can develop from it. Put simply, depending in which state (subpopulation) the toggle switch is, the cells will produce different fluorescent proteins upon addition of inducer molecules (ATc or IPTG). E.g. if ATc is added, one subpopulation will be red, while the other will be yellow although both carry exactly the same DNA information. Therefore the epigenetic feature here is the binding of specific repressor proteins, whose production is dependend on the toggle switch state, to their corresponding operator sequences.
System parts
educatETH E.coli consists of 11 parts that can be synthesized independently (want to know how this is done in the lab? Then visit our In the Lab page!) Please note that four of them (4,5 and 8,9) form together two functional system units. They have been separated to ensure comparable part lengths and thus enable easier introduction into plasmids.
| 1 | TetR production | [http://partsregistry.org/Part:BBa_I739001 BBa_I739001] | constitutive subsystem | |
|---|---|---|---|---|
| 2 | LacI production | [http://partsregistry.org/Part:BBa_I739002 BBa_I739002] | constitutive subsystem | |
| 3 | LuxR production | [http://partsregistry.org/Part:BBa_I739003 BBa_I739003] | constitutive subsystem | |
| 4 | 1st half of P22 cII / EYFP production | [http://partsregistry.org/Part:BBa_I739004 BBa_I739004] | reporting subsystem | |
| 5 | 2nd half of P22 cII / EYFP production | [http://partsregistry.org/Part:BBa_I739005 BBa_I739005] | reporting subsystem | |
| 6 | cI production | [http://partsregistry.org/Part:BBa_I739006 BBa_I739006] | learning subsystem | |
| 7 | P22 cII production | [http://partsregistry.org/Part:BBa_I739007 BBa_I739007] | learning subsystem | |
| 8 | 1st half of cI / ECFP production | [http://partsregistry.org/Part:BBa_I739008 BBa_I739008] | reporting subsystem | |
| 9 | 2nd half of cI / ECFP production | [http://partsregistry.org/Part:BBa_I739009 BBa_I739009] | reporting subsystem | |
| 10 | RFP production | [http://partsregistry.org/Part:BBa_I739010 BBa_I739010] | reporting subsystem | |
| 11 | GFP production | [http://partsregistry.org/Part:BBa_I739011 BBa_I739011] | reporting subsystem |
| 1+2+3 | tetR + lacI + luxR production | [http://partsregistry.org/Part:BBa_I739013 BBa_I739013] | constitutive subsystem | |
|---|---|---|---|---|
| 4+5 | P22 cII + EYFP production | [http://partsregistry.org/Part:BBa_I739015 BBa_I739015] | reporting subsystem | |
| 8+9 | cI + ECFP production | [http://partsregistry.org/Part:BBa_I739016 BBa_I739016] | reporting subsystem | |
| (4+5)+(8+9) | (P22 cII + EYFP) + (cI + ECFP) production | [http://partsregistry.org/Part:BBa_I739017 BBa_I739017] | reporting subsystem | |
| 6+7 | cI + P22 cII production | [http://partsregistry.org/Part:BBa_I739018 BBa_I739018] | learning subsystem | |
| 10+11 | RFP + GFP production | [http://partsregistry.org/Part:BBa_I739019 BBa_I739019] | reporting subsystem | |
| (6+7)+(10+11) | (cI + P22 cII) + (RFP + GFP) production | [http://partsregistry.org/Part:BBa_I739020 BBa_I739020] | learning/reporting subsystem |
| 1' | cI negative / tetR negative promoter | [http://partsregistry.org/Part:BBa_I739102 BBa_I739102] | reporting subsystem | |
|---|---|---|---|---|
| 2' | lacI negative / P22 cII negative promoter | [http://partsregistry.org/Part:BBa_I739103 BBa_I739103] | reporting subsystem | |
| 3' | luxR/HSL positive / P22 cII negative promoter | [http://partsregistry.org/Part:BBa_I739104 BBa_I739104] | learning subsystem | |
| 4' | luxR/HSL positive / cI negative promoter | [http://partsregistry.org/Part:BBa_I739105 BBa_I739105] | learning subsystem | |
| 5' | tetR negative / P22 cII negative promoter | [http://partsregistry.org/Part:BBa_I739106 BBa_I739106] | reporting subsystem | |
| 6' | cI negative / lacI negative promoter | [http://partsregistry.org/Part:BBa_I739107 BBa_I739107] | reporting subsystem |
| 1" | PoC promoter | [http://partsregistry.org/Part:BBa_I739101 BBa_I739101] | proof of concept, no part of the system | |
|---|---|---|---|---|
| 2" | PoC intermediate | [http://partsregistry.org/Part:BBa_I739014 BBa_I739014] | proof of concept, no part of the system | |
| 3" | PoC composite | [http://partsregistry.org/Part:BBa_I739021 BBa_I739021] | proof of concept, no part of the system |
Applications
Besides being a nice model for describing learning behavior or epigenetics, our system-structure can also be used for potent real world applications:
Multipurpose Cell Lines =
Our system can be trained to behave in a specific way by setting its inducible toggle switch to one of its two states. Let’s say this specific states trigger specific and different events such as enzyme synthesis, transcriptional regulation, virion production or cell death. In this case one can view the bacterial cell line containing this system as a multipurpose cell line. Put another way: One has a cell line and adds a certain chemical to train it to the desired behaviour instead of constructing two independent cell lines. This means, one applies an “input engineering” instead of a “DNA engineering” approach. If one extends this idea to several inducible toggle switches being harboured in the same cell line, the number of possible phenotypes increases to 2^n, whereas n equals the number of toggle switches. E.g. if one would have 5 toggle switches inside a cell line, 32 different behaviour patterns would be possible.
Intelligent Biosensors and self-adaptation
As a direct application of the multipurpose strains mentioned above we constructed a system capable of sensing different chemicals and producing different fluorescent proteins. Since the cells can be trained to produce one of several specific fluorescent proteins when a certain chemical is present, one can also view those cells as intelligent biosensors, able to adapt themselves to a certain environment. Alternatively those sensors could of course also be trained manually.
References
[http://www.nature.com/nature/journal/v403/n6767/abs/403335a0.html [1] Elowitz MB and Leibler S] "A synthetic oscillatory network of transcriptional regulators", Nature 403:335-338, 2000
[http://www.nature.com/nature/journal/v447/n7143/abs/nature05913.html;jsessionid=62903C604764B175945C03DB8639ECBD [2] Bird A] "Perceptions of epigenetics", Nature 447:396-398, 2007
[http://linkinghub.elsevier.com/retrieve/pii/S096098220701007X [3] Ptashne M] "On the use of the word ‘epigenetic’", Current Biology 17(7):R233-R236, 2007
To Do
New
<p>- Update and correct parts in parts list. Write better in a table
- Update and correct full system scheme
- What is the proof of concept mentioned?
- Are you sure about the reporters? This is not what I understood. I though YFP and CFP were also during testing. Yes! I'm still updating the text :-)
- I don't think it is wise to make p22cII in P22 cII, since we have in the first way all over the place. Stefan: Yes I know :-( However we have different descriptions: p22cII, cII, P22CII, p22 cII, P22 cII. However, P22 cII is the correct one. I'm not sure how we can solve this (minor!) problem.
- Try to improve table with system phases. It doesn't look so nice...
- In the memorizing phase, is there color or not? - Nope, there is no color - There are colors, however they will disappear within 2-3 hours.
- We need to put the complete spaghetti system as well.
- Check my terminology (operator sites etc)
- Put Stefan's updated part on epigenetics
- Fill in table completely, make it more reading-friendly