Glasgow/Modeling
From 2007.igem.org
(→Trial model for the PDntR|S promoter) |
(→'''BioBrick® library''') |
||
| Line 52: | Line 52: | ||
| - | == ''' | + | == '''BioBrick library''' == |
[http://compbio.dcs.gla.ac.uk/iGEM2007/BioBricklibrary.zip BioBrickLibrary.zip] | [http://compbio.dcs.gla.ac.uk/iGEM2007/BioBricklibrary.zip BioBrickLibrary.zip] | ||
An add on Library to Simulink for modelling dynamical biological systems at Brick (Gene) level. Simulink is a program dedicated for dynamical system simulation, however in depth knowledge of dynamics is needed if one is to simulate system mentioned above. The BioBrick® library has all the blocks as well as GUI (Graphical User Interface) needed to do the job without understanding how Simulink works. It uses drag and drop system and shares all constants in Matlab’s m file, so it is easy to store and update them.<br> | An add on Library to Simulink for modelling dynamical biological systems at Brick (Gene) level. Simulink is a program dedicated for dynamical system simulation, however in depth knowledge of dynamics is needed if one is to simulate system mentioned above. The BioBrick® library has all the blocks as well as GUI (Graphical User Interface) needed to do the job without understanding how Simulink works. It uses drag and drop system and shares all constants in Matlab’s m file, so it is easy to store and update them.<br> | ||
Revision as of 14:23, 2 October 2007
| https://static.igem.org/mediawiki/2007/thumb/c/cc/Uog.jpg/50px-Uog.jpg | Back To Glasgow's Main Page |
|---|
Trial model for the PDntR|S promoter
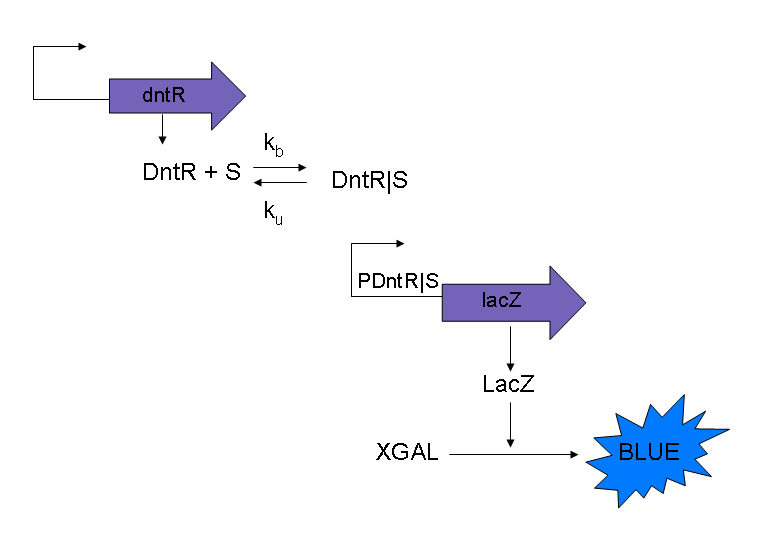
The current model that will be used for modeling purposes will use the three following equations to model the simple system as shown in Figure 1. The reasons for using these modified equations are discussed below.
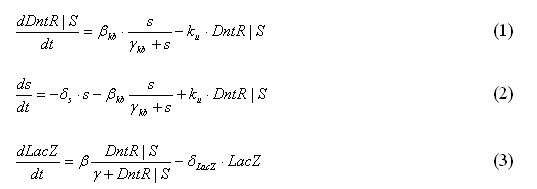
Discussion
The equation for the change of DntR|S within the system could be modelled by Equation 4
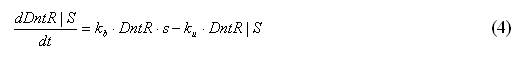
At the beginning of the reaction DntR will have an unknown constant value as it cannot be measured physically in the laboratory. As the DntR will have a constant value the DntR and kb can be replaced by another constant kb’ as shown
By substituting Equation 5 into Equation 4 gives

However it is clear from Equation 6 that as the signal increases the DntR|S will increase without ever reaching a saturation limit. It was decided that the equation should be changed to utilize Michaelis-Menten. This means that a saturation limit will now be possible as shown in Equation 1.
Next it is important to note that the signal equation includes a term for the degradation of the signal as this is possible in the physical model. This equation also includes a term for the binding and a term for the unbinding as shown in Equation 2.
Also it should be noted that in this model XGAL is ignored, this is because the level of blue that is measured is proportional to the LacZ produced. By ignoring XGAL it will simplify the system equations.
Another point to note is that in this model of the system the transcription and the translation steps have been ‘lumped’ together by using Michaelis-Menten rather than modeling these biological steps separately.
Minicap Sensitivity Analysis Program Package Abstract
The Multi-Parametric and Initial Concentration Sensitivity analysis (Minicap) package is a Matlab function which executes a chosen Dynamic or Stochastic System Function for a defined number of different variable values across any desired range. The subject of analysis can either be constants in the user's system eg. parameters in a biological system (MPSA) or initial values of the variables in the user's system eg. initial substrate concentrations (ISCSA). The program will output a plot for each variable showing a comparison of acceptable and unacceptable samples across the subject's range with 3 calculated quantitative comparison figures: the Correlation Coefficient, the Area between acceptable and unacceptable curves, and the Standard Deviation of the gradient of the acceptable plot. The comparative and intrinsic sensitivity of each chosen subject is thus highlighted. A plot showing the trend of the Substrate of Interest over time is also displayed.
As well as this report, the Minicap package contains a User Manual in html, a number of example codes and all the novel (i.e not ode15s) function and text files required to run Minicap.
To see the full PDF report on Minicap [click here].
BioBrick library
[http://compbio.dcs.gla.ac.uk/iGEM2007/BioBricklibrary.zip BioBrickLibrary.zip]
An add on Library to Simulink for modelling dynamical biological systems at Brick (Gene) level. Simulink is a program dedicated for dynamical system simulation, however in depth knowledge of dynamics is needed if one is to simulate system mentioned above. The BioBrick® library has all the blocks as well as GUI (Graphical User Interface) needed to do the job without understanding how Simulink works. It uses drag and drop system and shares all constants in Matlab’s m file, so it is easy to store and update them.
BioBrick® library’s main aim is to tell whether and how different topology will influence the output of the system. If actual rate constants are known it can be used instead or as complimentary to ODE modelling. However it must be noted that ODE rate constants ARE NOT TRANFERABLE to BioBrick ® library.
More information can be found in :ElectrEcoBluSimulinkManual