Glasgow/Plan
From 2007.igem.org
| Line 9: | Line 9: | ||
<center>[[Image:ElectrEcoBlu.jpg|frame]]</center> | <center>[[Image:ElectrEcoBlu.jpg|frame]]</center> | ||
<br> | <br> | ||
| - | ElectrEcoBlu combines an environmental biosensor for common organic pollutants with a microbial fuel cell which can produce its own electricity. | + | <font face=georgia color=#3366CC size=4>ElectrEcoBlu</font> |
| + | <br> | ||
| + | ElectrEcoBlu combines an environmental biosensor for common organic pollutants with a microbial fuel cell which can produce its own electricity. These cells produce their own electrical power output which increases in the presence of one or more organic pollutant stimulants. This system has the potential to be used for self-powered long term ''in situ'' and online monitoring with an electrical readout. It is based around novel reporter genes encoding electron carrying mediators which aid the transfer of electrons from the cells to the electrodes resulting in enhanced electricity generation. | ||
<br> | <br> | ||
<br> | <br> | ||
'''Environmental Biosensor''' | '''Environmental Biosensor''' | ||
<br> | <br> | ||
| - | + | ||
| + | A biosensor is “a self contained integrated device consisting of a biological recognition element (enzyme, antibody, receptor or microorganism) which is interfaced to a chemical sensor (i.e., analytical device) that together reversibly respond in a concentration-dependent manner to a chemical species”. Biosensors offer advantages over current analytical methods for environmental applications such as the possibility of portability and working on-site (Paitan et al, 2003), and being mre cost- and time-effective with the ability of measuring pollutants in complex matrices with minimal sample preparation Ron, 2007). Microorganisms are being developed to exhibit a quick, detectable response to low levels of contamination. These biosensors have the potential to be maintained on-site where they can monitor conditions constantly. For example, biosensors could be used in the soil or water outside of industrial factories to ensure that discharge from the factory is acceptable at all times, or at nuclear reactor sites to make ensure that radioactive materials are not being released into the environment. | ||
| + | |||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 18:03, 8 October 2007
| https://static.igem.org/mediawiki/2007/thumb/c/cc/Uog.jpg/50px-Uog.jpg | Back To Glasgow's Main Page |
|---|
ElectrEcoBlu
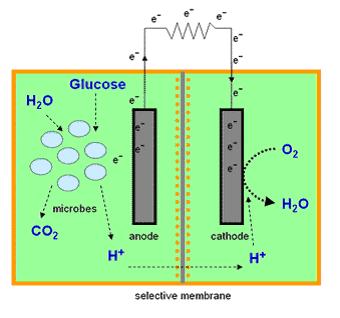
ElectrEcoBlu combines an environmental biosensor for common organic pollutants with a microbial fuel cell which can produce its own electricity. These cells produce their own electrical power output which increases in the presence of one or more organic pollutant stimulants. This system has the potential to be used for self-powered long term in situ and online monitoring with an electrical readout. It is based around novel reporter genes encoding electron carrying mediators which aid the transfer of electrons from the cells to the electrodes resulting in enhanced electricity generation.
Environmental Biosensor
A biosensor is “a self contained integrated device consisting of a biological recognition element (enzyme, antibody, receptor or microorganism) which is interfaced to a chemical sensor (i.e., analytical device) that together reversibly respond in a concentration-dependent manner to a chemical species”. Biosensors offer advantages over current analytical methods for environmental applications such as the possibility of portability and working on-site (Paitan et al, 2003), and being mre cost- and time-effective with the ability of measuring pollutants in complex matrices with minimal sample preparation Ron, 2007). Microorganisms are being developed to exhibit a quick, detectable response to low levels of contamination. These biosensors have the potential to be maintained on-site where they can monitor conditions constantly. For example, biosensors could be used in the soil or water outside of industrial factories to ensure that discharge from the factory is acceptable at all times, or at nuclear reactor sites to make ensure that radioactive materials are not being released into the environment.
Microbial Fuel Cells
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Novel Reporter System
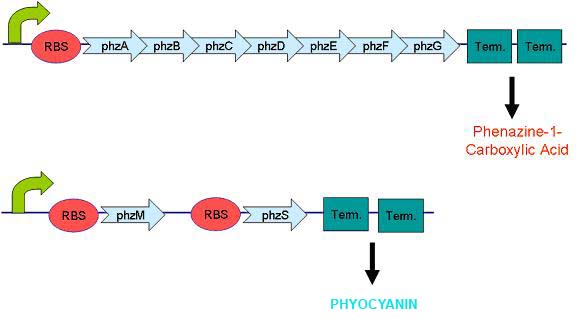
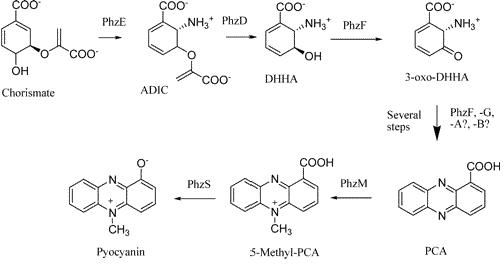
Pyocyanin is the main phenazine compound produced by Pseudomonas aeruginosa as a result of quorum sensing. Pyocyanin engages in oxidation-reduction reactions which deplete cells of NADH, glutathione, and other antioxidants and produces oxidants such as superoxide and peroxides (Parsons, 2006). By using the genes phzM, phzS and the seven gene operon phzABCDEFG which express pyocyanin in P. aeruginosa, we intend to harness the oxidation-reduction potential of pyocyanin to power a microbial fuel cell.
XylR and BTEX Chemicals
Benzene, Toluene, Ethylbenzene and Xylenes (BTEX chemicals) are types of volatile organic hydrocarbons found in petroleum derivatives. Contamination of BTEX chemicals, which can be toxic and carcinogens, is typically located near to petroleum and natural gas production sites, and in areas with ground storage tanks.
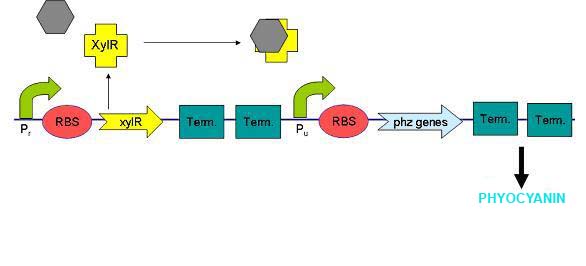
XylR protein, which has evolved in water- and soil-borne bacteria, alters in shape when bound to toluene-like chemicals and forms a transcriptional activator which binds to promoter Pu triggering a cascade of metabolic reactions (Willardson et al, 1998). We intend to use xylR and related promoters Pr and Pu to produce pyocyanin in the presence of toluene.
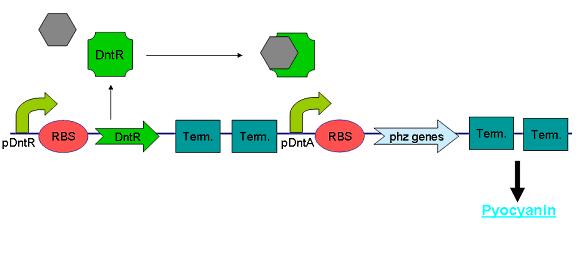
DntR and Dinitrotoluenes
Dinitrotoluene (DNT) is an intermediate in the commercial production of TNT and is often found as an environmental contaminant in areas surrounding production sites. Being toxic and carcinogenic, it is a pollutant of major concern and therefore previous works have been undertaken to characterize the DNT recognition and degradation operons of Burkholderia cepacia strain R34 – an isolate found to be able to utilise 2,4-DNT as its sole carbon source (R.J. Spanggord 1991). During the course of our project we attempted to sub-clone the initial regulatory part of these pathways from a Pseudomonas strain carrying the relevant operon (Rosser & Bruce) with the intention of using it as the sensing component of our biosensor.
The regulatory region consists of a putative LysR-type transcription factor (DntR) consisting of an N-terminal DNA-binding domain and a C-terminal cofactor-binding domain, 301 amino acids in length (Johnson 2002) which recognises 2,4-DNT and positively regulates transcription of an operon encoding components of the 2,4-DNT dioxygenase complex. We designed primers to the 3’ end of DntR and to the region immediately upstream of the first ORF in the operon (see Fig X, below) resulting in a product that conveniently contained both DntR and its target promoter.