Glasgow/Wetlab/Results
From 2007.igem.org
(→Results) |
(→Results) |
||
| Line 13: | Line 13: | ||
The XylR transcriptional regulator system was initially tested through the induction of the reporter gene beta-galactosidase, linked to the pU promoter. Initial studies were not performed in a biobrick system as they were occurring concurrently with the creation of novel biobricks. Instead a construct was utilised based on the pQF52 reporter plasmid where the xylA promoter was fused to the lacZ reporter gene. The xylR regulatory gene was cloned in the opposite orientation. The plasmid was termed pQF52 xylR.<br> | The XylR transcriptional regulator system was initially tested through the induction of the reporter gene beta-galactosidase, linked to the pU promoter. Initial studies were not performed in a biobrick system as they were occurring concurrently with the creation of novel biobricks. Instead a construct was utilised based on the pQF52 reporter plasmid where the xylA promoter was fused to the lacZ reporter gene. The xylR regulatory gene was cloned in the opposite orientation. The plasmid was termed pQF52 xylR.<br> | ||
<br> | <br> | ||
| - | E.coli containing pQF52 dntR were grown in LB culture overnight, before addition of a range of aromatic environmental pollutants were added. Production of lacZ was assayed for by the Miller Assay ([[Glasgow/Wetlab/Protocols#Protocol 10: Miller Assay|Protocol 10]]). | + | E.coli containing pQF52 dntR were grown in LB culture overnight, before addition of a range of aromatic environmental pollutants were added. Production of lacZ was assayed for by the Miller Assay ([[Glasgow/Wetlab/Protocols#Protocol 10: Miller Assay|Protocol 10]]).<br> |
| + | <br> | ||
| + | As seen in Figure 1, lacZ production increased signicantly as salicylate concentration increased. A low background production of lacZ was observed in the absence of environmental pollutant. | ||
| + | |||
| + | [[Image:Graph1.PNG|frame]] | ||
Revision as of 17:18, 25 October 2007
 | Back To Glasgow's Main Page | Back To Glasgow's Wetlab Log | Back To Glasgow's Project Page |
|---|
Results
Pyocyanin
The genes encoding the two final enzymes involved in the pyocyanin biosynthesis pathway, Phz M and Phz S, were cloned and successfully created into biobricks. They have been subcloned to include terminators after the genes. The phenzine biosynthesis operon responsible for the production of the precursor phenazine PCA, was over 7kb long and proved troublesome to clone. An alternative strategy involved cloning this operon in two parts. PhzE, PhzF and PhzG were cloned into biobrick format and are currently undergoing site directed mutagenesis. Pseudomonas fluorescens 2-71 was obtained as an alternative source of PCA.
XylR and BETX Chemicals
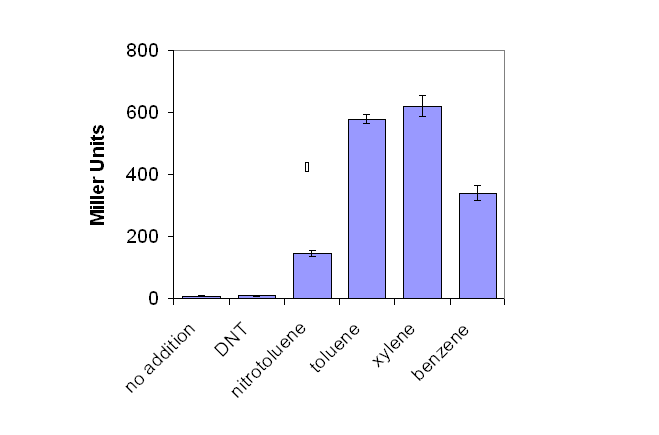
The XylR transcriptional regulator system was initially tested through the induction of the reporter gene beta-galactosidase, linked to the pU promoter. Initial studies were not performed in a biobrick system as they were occurring concurrently with the creation of novel biobricks. Instead a construct was utilised based on the pQF52 reporter plasmid where the xylA promoter was fused to the lacZ reporter gene. The xylR regulatory gene was cloned in the opposite orientation. The plasmid was termed pQF52 xylR.
E.coli containing pQF52 dntR were grown in LB culture overnight, before addition of a range of aromatic environmental pollutants were added. Production of lacZ was assayed for by the Miller Assay (Protocol 10).
As seen in Figure 1, lacZ production increased signicantly as salicylate concentration increased. A low background production of lacZ was observed in the absence of environmental pollutant.