Glasgow/Wetlab/Results
From 2007.igem.org
(→Results) |
|||
| Line 8: | Line 8: | ||
<font face=georgia color=#0000FF size=4>Pyocyanin</font><br> | <font face=georgia color=#0000FF size=4>Pyocyanin</font><br> | ||
| - | The genes encoding the two final enzymes involved in the pyocyanin biosynthesis pathway, | + | The genes encoding the two final enzymes involved in the pyocyanin biosynthesis pathway, PhzM and PhzS, were cloned and successfully made into BioBricks. They have been subcloned to include terminators after the genes. The phenazine biosynthesis operon responsible for the production of the pyocyanin precursor phenazine-1-carboxylic acid (PCA) was over 7kb long and proved troublesome to clone. An alternative strategy involved cloning this operon in two parts. PhzE, PhzF and PhzG were cloned into BioBrick format and are currently undergoing site directed mutagenesis. ''Pseudomonas fluorescens'' 2-71 was obtained as an alternative source of PCA.<br> |
<br> | <br> | ||
<font face=georgia color=#0000FF size=4>XylR and BETX Chemicals</font><br> | <font face=georgia color=#0000FF size=4>XylR and BETX Chemicals</font><br> | ||
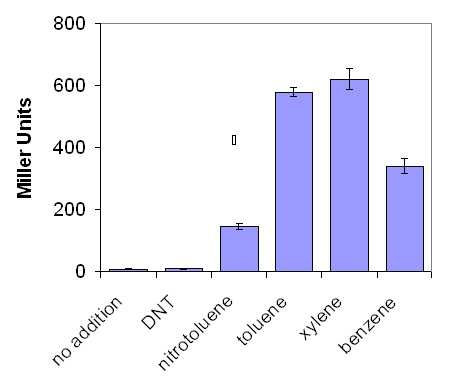
| - | <center>[[Image:Graph1b.PNG|frame|'''Figure 1:''' The effect of a variety of environmental pollutants on lacZ production by E.coli containing pQF52 xylR, when grown on LB media, as measured by the Miller Assay.]]</center> | + | <center>[[Image:Graph1b.PNG|frame|'''Figure 1:''' The effect of a variety of environmental pollutants on lacZ production by ''E. coli'' containing pQF52 xylR, when grown on LB media, as measured by the Miller Assay.]]</center> |
<br> | <br> | ||
| - | The XylR transcriptional regulator system was initially tested | + | The XylR transcriptional regulator system was initially tested for the induction of the reporter gene lacZ driven by the Pu promoter. Initial studies were not performed in a BioBrick system as they were occurring in parallel with the creation of novel BioBricks. Instead, a construct based on the pQF52 reporter plasmid containing a XylA:lacZ fusion reporter gene was utilised (XylA is also activated by xylene-bound XylR). The XylR regulatory gene was cloned in the opposite orientation. The plasmid was termed pQF52 XylR '''''(see Fig X)'''''.<br> |
<br> | <br> | ||
| - | + | '''''Have we got this part right? Was XylR in pQF52? Because it says below that DntR was also in pQF52, so surely one of them must be the wrong way round.''''' [[User:Scott.w.ramsay|Scott.w.ramsay]] 06:25, 26 October 2007 (EDT) | |
<br> | <br> | ||
| - | + | <br> | |
| + | ''E. coli'' containing pQF52 XylR were grown in LB culture overnight before addition of a range of aromatic environmental pollutants were added. Production of lacZ was assayed for by the Miller Assay ([[Glasgow/Wetlab/Protocols#Protocol 10: Miller Assay|Protocol 10]]).<br> | ||
| + | <br> | ||
| + | LacZ production was greatest in response to xylene but also reached comparable levels when the bacteria were challenged with benzene and toluene (figure 1). This would be of benefit to an environmental system where the aim is purely to detect the incidence of pollution, reducing the materials and costs associated with sensing as many different chemicals as possible. XylR was not deemed sensitive enough to nitrotoluene and dinitrotoluene (DNT) to do any further investigation with those pollutants.<br> | ||
| - | The XylR system was considered suitable for further development and so | + | The XylR system was considered suitable for further development and so BioBricks were constructed to facilitate a biosensor that produces pyocyanin. In order to test alternative reporter systems, a sensor system where XylR transcriptionally regulates the production of the Renilla luciferase has been successfully created using the RBS [http://partsregistry.org/Part:BBa_J61101 BBa_J61101] and the luciferase cds [http://partsregistry.org/Part:BBa_J52008 BBa_J52008].<br> |
<br> | <br> | ||
<font face=georgia color=#0000FF size=4>DntR and Dinitrotoluenes</font><br> | <font face=georgia color=#0000FF size=4>DntR and Dinitrotoluenes</font><br> | ||
Revision as of 10:25, 26 October 2007
 | Back To Glasgow's Main Page | Back To Glasgow's Wetlab Log | Back To Glasgow's Project Page |
|---|
Results
Pyocyanin
The genes encoding the two final enzymes involved in the pyocyanin biosynthesis pathway, PhzM and PhzS, were cloned and successfully made into BioBricks. They have been subcloned to include terminators after the genes. The phenazine biosynthesis operon responsible for the production of the pyocyanin precursor phenazine-1-carboxylic acid (PCA) was over 7kb long and proved troublesome to clone. An alternative strategy involved cloning this operon in two parts. PhzE, PhzF and PhzG were cloned into BioBrick format and are currently undergoing site directed mutagenesis. Pseudomonas fluorescens 2-71 was obtained as an alternative source of PCA.
XylR and BETX Chemicals
The XylR transcriptional regulator system was initially tested for the induction of the reporter gene lacZ driven by the Pu promoter. Initial studies were not performed in a BioBrick system as they were occurring in parallel with the creation of novel BioBricks. Instead, a construct based on the pQF52 reporter plasmid containing a XylA:lacZ fusion reporter gene was utilised (XylA is also activated by xylene-bound XylR). The XylR regulatory gene was cloned in the opposite orientation. The plasmid was termed pQF52 XylR (see Fig X).
Have we got this part right? Was XylR in pQF52? Because it says below that DntR was also in pQF52, so surely one of them must be the wrong way round. Scott.w.ramsay 06:25, 26 October 2007 (EDT)
E. coli containing pQF52 XylR were grown in LB culture overnight before addition of a range of aromatic environmental pollutants were added. Production of lacZ was assayed for by the Miller Assay (Protocol 10).
LacZ production was greatest in response to xylene but also reached comparable levels when the bacteria were challenged with benzene and toluene (figure 1). This would be of benefit to an environmental system where the aim is purely to detect the incidence of pollution, reducing the materials and costs associated with sensing as many different chemicals as possible. XylR was not deemed sensitive enough to nitrotoluene and dinitrotoluene (DNT) to do any further investigation with those pollutants.
The XylR system was considered suitable for further development and so BioBricks were constructed to facilitate a biosensor that produces pyocyanin. In order to test alternative reporter systems, a sensor system where XylR transcriptionally regulates the production of the Renilla luciferase has been successfully created using the RBS BBa_J61101 and the luciferase cds BBa_J52008.
DntR and Dinitrotoluenes
The DntR transcriptional regulator system was tested through the induction of the reporter gene beta-galactosidase, linked to the dntA promoter. Initial studies were not performed in a biobrick system as they were occurring concurrently with the creation of novel biobricks. Instead a construct was utilised based on the pQF52 reporter plasmid where the dntA promoter was fused to the lacZ reporter gene. The dmpR regulatory gene was cloned in the opposite orientation. The plasmid was termed pQF52 dntR.
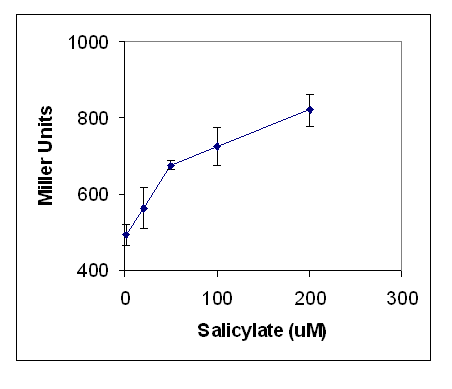
E.coli containing pQF52 dntR were grown in LB culture overnight, before addition of a range of concentrations of salicylate were added. Production of lacZ was assayed for by the Miller Assay (Protocol 10).
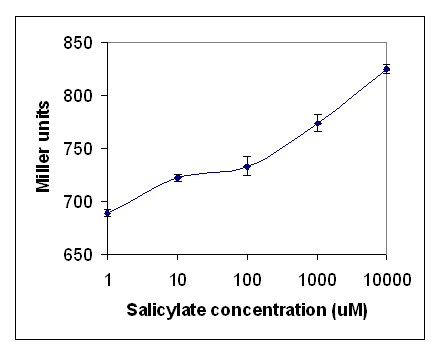
As seen in Figures 2 and 3, lacZ production increased signicantly as salicylate concentration increased. A background production of lacZ was observed, therefore further testing was performed to optimise the assay by growing the cells on minimal media. The reason for this was to limit the amounts of tryptophan which was considered a potential mimic of salicylate. The range of concentrations of salicylate were extended to determine the linearity of the response. The results are shown in Figure X. There was no reduction in background activity with no salicylate. As salicylate increased the response increased, this shows that the sensitivity of the assay is from 10 um to 10 mM.
The dntR system was considered suitable for further development and so biobricks were constructed to facilitate a biosensor involving the production of the electrochemical mediator pyocyanin.
DmpR and Phenolic Compounds
The DmpR transcriptional regulator system was tested through the induction of the reporter gene beta-galactosidase, linked to the dmp operon promoter. Initial studies were not performed in a biobrick system as they were occurring concurrently with the creation of novel biobricks. Instead a construct was obtained by personal communication with A.Wise where the promoter of the dmp operon had been fused to the lacZ reporter gene and inserted chromosomally at the trp locus. The dmpR regulatory gene was contained on a plasmid – p322-dmpR (RI) which was a derivative of pBR322.
E.coli containing the chromosomal insert and the plasmid p322-dmpR (RI) were grown on a variety of agar media. The media that were tested were rich media (Luria-Bertani - LB), minimal media and spent LB media. The colonies were blotted onto a sheet of hybrid N nylon membrane. The membrane was placed in a petri dish on top of thin layer of agar containing X-gal before adding phenol, cresols, dinitrophenols, dimethylphenols and dichlorophenols and incubated for 2 hrs at 37 C. The membranes were then assessed for the production of blue colouration.
There was shown to be a high background production of lacZ in all of the media tried. The spent LB was the best of the media tried, althought even with this there was a high background level of lacZ production. This was thought to be due to the presence of the amino acid, tryptophan, in the media. This amino acid is essential for the growth of E.coli. Its chemical structure is similar to phenol and could therefore be triggering expression of lacZ.
It was decided that due to the background induction of lacZ by tryptophan, the transcriptional regulator DmpR would not be studied further. The production of the relevant biobricks was therefore halted.