Imperial/Wet Lab/Protocols/CBD2.2
From 2007.igem.org
(Difference between revisions)
m |
m |
||
| Line 1: | Line 1: | ||
{{Template: IC07navmenu}} | {{Template: IC07navmenu}} | ||
| + | <br clear="all"> | ||
__NOTOC__ | __NOTOC__ | ||
Revision as of 02:23, 27 October 2007

Wet Lab: Protocols: Isothermal experiments with pTet-GFPmut3b construct
Aims:
- To measure the fluorescence over a 6 hour period, for a range of temperatures.
- To continue from the temperatures measured before, 37oC and 20oC; temperatures 8oC, 15oC, 25oC and 30oC are measured this time round.
- 8oC is the hypothesized minimum temperature where the system stops working. Judging from the behaviour of the system at this temperature, we will refine its working parameters.
Equipment
- Water Baths at in cold room at 8°C/15°C and in the open at 25°C/30°C
- Fluorometer + PC
- 4 Fluorometer plates (black)
- Sticky seal tape
- Gilson pipettes p1000 p200 p20 p10
- Eppendorf Tubes
- Stopwatch
Reagents
- Commercial S30 E.coli extract. Including:
- 175µl Amino Acid Mixture Minus Cysteine, 1mM
- 175µl Amino Acid Mixture Minus Methionine, 1mM
- 175µl Amino Acid Mixture Minus Leucine, 1mM
- 450µl S30 Extract, Circular (3 × 150µl)
- 750µl S30 Premix Without Amino Acids
- DNA pTet-GFP from maxiprep
- Nuclease Free water
- GFP solution
Protocol
- First collect all equipment and reagents and ensure that the fluorometer and that the PC connected has a data collection protocol installed.
- Place each of the 96 well plates in their respective incubators so as to heat them up to the appropriate temperature before the experiments start.
- For Each Temperature Carry out the following Procedure
- Commercial E.coli Cell Extract: Add 240ul of cell extract + 320ul of premix + 40ul of amino acid mix minus cys + 40ul amino acid mix minus leu to an eppendorf tube, labelled A.
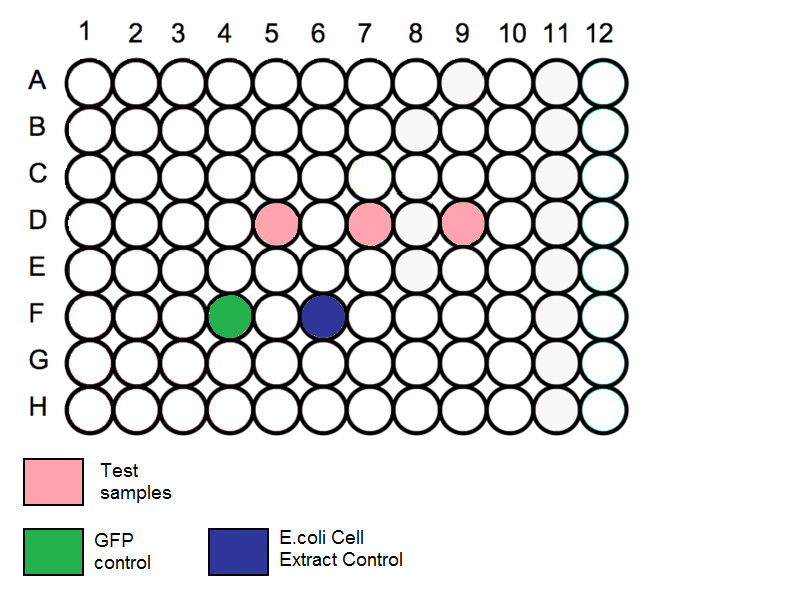
Loading Plate
- Add 60ul of water to all the outermost wells of the plate to minimise evaporation of the samples.
- Add 40ul of cell extract preparation from A to 4 wells each, on a 96 well plate. (The 4 wells should be placed towards the middle of the plate)
- Add 2ug of DNA each to 3 of the wells to make up to 60ul in each well.
- In the last well, add 20ul of nuclease free water to the cell extract preparation.
- Place the plate in the fluorometer to measure its initial fluorescent reading.
- After the measurement, place the sticky tape across the plate, and put the plate in the 8oC water bath.
- Start on the next plate, and repeat procedures 2-6.
- Place the plate in the 15oC water bath.
- Repeat the same procedures for the remaining two plates, except that before placing them in the water bath, wrap aluminium foil around them to prevent photobleaching. (This is because the water baths 25oC and 30oC will be out in the openand exposed to light)
- Stagger the start of all the plates by around 5 minutes, and measure the temperature every 30 minutes for each temperature.
- Continue this for 6 hours, and plot a graph of the fluorescence against time for all 4 plates.
|