Mechanism II
From 2007.igem.org
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
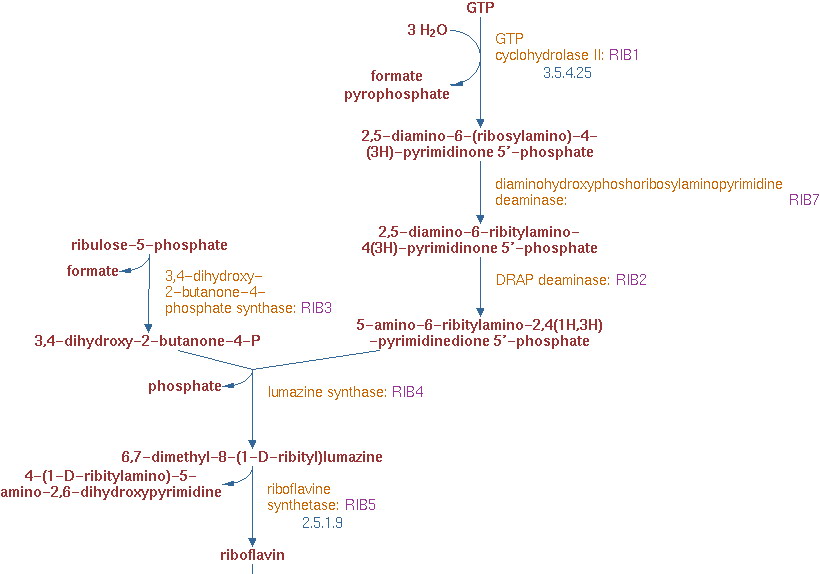
| - | Yeast can produce riboflavin itself, but it can only supply its own metabolism without accumulation. There are 6 genes related to the riboflavin production. The | + | Yeast can produce riboflavin itself, but it can only supply its own metabolism without accumulation. There are 6 genes related to the riboflavin production. The pathway of riboflavin synthesis is showed below: |
[[Image:TJU_TEAM2_MIIFiguer4.jpg|thumb|600px|center|'''Figure 4 the riboflavin synthesis pathway''']] | [[Image:TJU_TEAM2_MIIFiguer4.jpg|thumb|600px|center|'''Figure 4 the riboflavin synthesis pathway''']] | ||
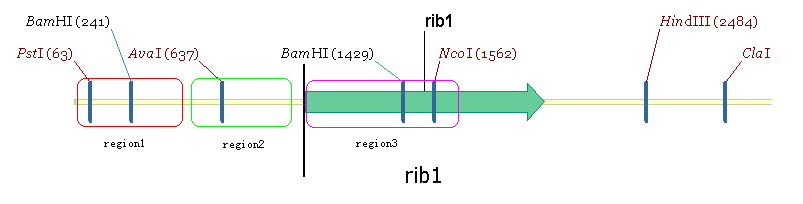
We propose to change the prmoters related to riboflavin synthesis of Yeast to strong promter ADH1. The two-step-gene replacement method is used in our project. Take RIB1 as an example. Take the RIB1gene as three regions: region1, region2 and region2 (figure5) | We propose to change the prmoters related to riboflavin synthesis of Yeast to strong promter ADH1. The two-step-gene replacement method is used in our project. Take RIB1 as an example. Take the RIB1gene as three regions: region1, region2 and region2 (figure5) | ||
| + | [[Image:TJU_TEAM2_MIIFiguer5.jpg|thumb|600px|center|'''Figure 5 the regions in RIB1 gene''']] | ||
| + | [[Image:TJU_TEAM2_MIIFiguer6.jpg|thumb|300px|right|'''Figure 6 plasmid Yiplac211''']] | ||
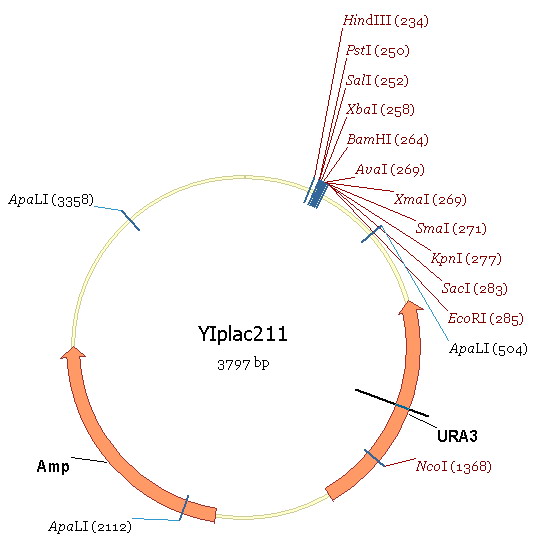
| + | Region 2 contains the RIB1 original promoter. Region3 is a part of gene ecoding region and region 1 is the upstream region of RIBI original promoter. Our aim is to substitude ADH1 for region2. We take the ADH1 strong promoter as region4. The plasmid used in this experiment is YIPlac211(figure 6). | ||
| + | The experimental process is: | ||
| + | #Digest the ADH1 promoter on puc18-ADH1 p+t with XbaI and BamHI, and then link it to YIPlac211. | ||
| + | #Design a primer and amplify the region3 from Yeast genome. The restriction site of up- and down-primer are SmaI and kpnI, respectively. Digest the amplified fragment with SmaI and kpnI, and then link it to the plasmid we have got in the first step. | ||
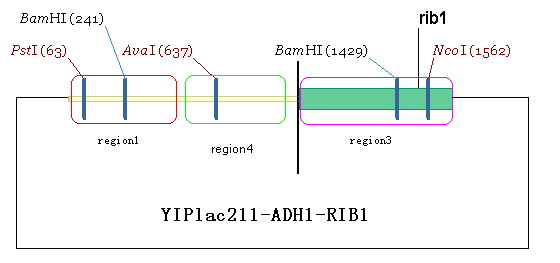
| + | #Design a primer and amplify the region1 from Yeast genome. The restriction site of up- and down-primer are HindIII and salI, respectively. Digest the amplified fragment with HindIII and salI, and then link it to the plasmid we have got in the second step. By doing this, we obtain plasmid YIPlac211-ADH1-RIB1(figure 7). | ||
| + | #Digest the construced plasmid we have got in step3 with PstI and transfer it into Yeast. The inherit maker is Ura3 and it can grow on the CM plate without uracil. Test the positive clone with PCR. | ||
| + | #Cultivate the positive colon on 5-FOA medium. The marker can spring out through homologous recombination. Filtrate the mutant we want with PCR. | ||
| + | For RIB1 and RIB3 are the key enzymes in riboflavin production, we operate on them in advance of other genes. | ||
| + | [[Image:TJU_TEAM2_MIIFiguer7.jpg|thumb|400px|center|'''Figure7 plasmid YIPlac211=ADH1-RIB1''']] | ||
Latest revision as of 06:04, 27 August 2007
Yeast can produce riboflavin itself, but it can only supply its own metabolism without accumulation. There are 6 genes related to the riboflavin production. The pathway of riboflavin synthesis is showed below:
We propose to change the prmoters related to riboflavin synthesis of Yeast to strong promter ADH1. The two-step-gene replacement method is used in our project. Take RIB1 as an example. Take the RIB1gene as three regions: region1, region2 and region2 (figure5)
Region 2 contains the RIB1 original promoter. Region3 is a part of gene ecoding region and region 1 is the upstream region of RIBI original promoter. Our aim is to substitude ADH1 for region2. We take the ADH1 strong promoter as region4. The plasmid used in this experiment is YIPlac211(figure 6). The experimental process is:
- Digest the ADH1 promoter on puc18-ADH1 p+t with XbaI and BamHI, and then link it to YIPlac211.
- Design a primer and amplify the region3 from Yeast genome. The restriction site of up- and down-primer are SmaI and kpnI, respectively. Digest the amplified fragment with SmaI and kpnI, and then link it to the plasmid we have got in the first step.
- Design a primer and amplify the region1 from Yeast genome. The restriction site of up- and down-primer are HindIII and salI, respectively. Digest the amplified fragment with HindIII and salI, and then link it to the plasmid we have got in the second step. By doing this, we obtain plasmid YIPlac211-ADH1-RIB1(figure 7).
- Digest the construced plasmid we have got in step3 with PstI and transfer it into Yeast. The inherit maker is Ura3 and it can grow on the CM plate without uracil. Test the positive clone with PCR.
- Cultivate the positive colon on 5-FOA medium. The marker can spring out through homologous recombination. Filtrate the mutant we want with PCR.
For RIB1 and RIB3 are the key enzymes in riboflavin production, we operate on them in advance of other genes.