Melbourne/Lab GV Notebook
From 2007.igem.org
(→PCR based insertion of Biobrick prefix and suffix, digestion and ligation to BBa_J61035) |
|||
| Line 18: | Line 18: | ||
The surrounding GC pairs come from the GC clamp we put on the end of the suffix, and the NotI site in the middle of the suffix. This point is worth noting on the suffix design page with a suggestion that AT pairs should be added between the suffix and GC clamp.Due to purchasing of a large number of primers that we didn't want to waste we persisted, swapping to cutting with the more central XbaI and SpeI and accepting the 50% drop in efficiency due to orientation uncertainty. The vector was cut with SpeI and treated with antarctic phosphatase. | The surrounding GC pairs come from the GC clamp we put on the end of the suffix, and the NotI site in the middle of the suffix. This point is worth noting on the suffix design page with a suggestion that AT pairs should be added between the suffix and GC clamp.Due to purchasing of a large number of primers that we didn't want to waste we persisted, swapping to cutting with the more central XbaI and SpeI and accepting the 50% drop in efficiency due to orientation uncertainty. The vector was cut with SpeI and treated with antarctic phosphatase. | ||
| - | + | [[Melbourne/Lab Notebook gv 6|Details Here]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
====Testing Floatation==== | ====Testing Floatation==== | ||
*Details: | *Details: | ||
Revision as of 12:27, 26 October 2007
<Return to Lab notebook> <team home page>
Contents |
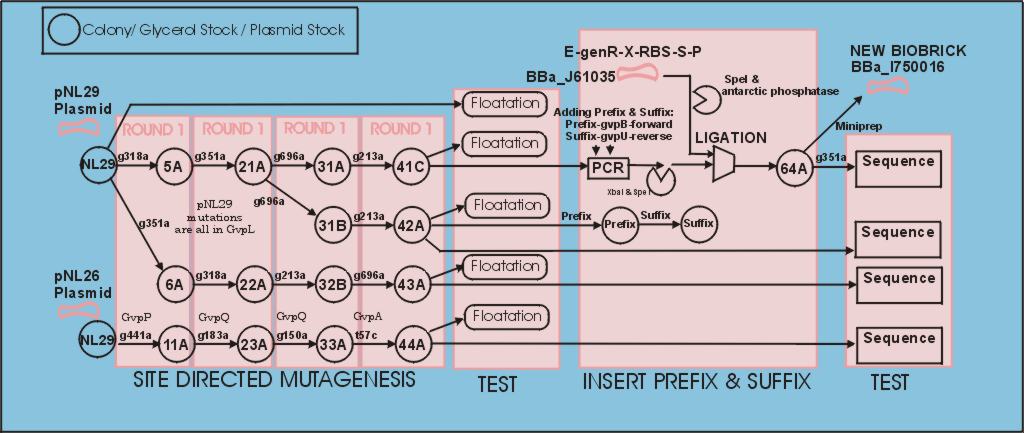
Developement overview (showing tube numbers)
=Steps:=Create Glycerol stocks of gas vesicle plasmids from Maura Cannon
Site dirrected mutagenesis:
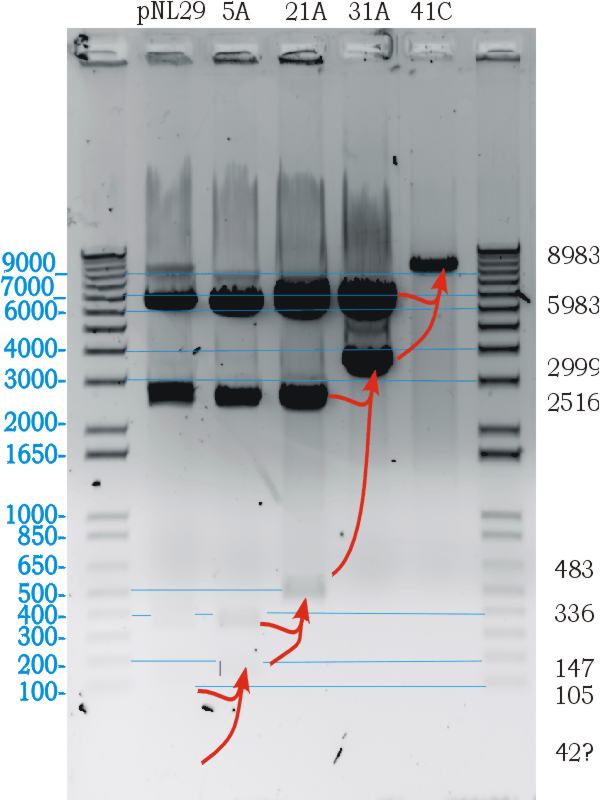
The polycistronic gas vesicle gene was obtained in an expression plasmid from Maura Cannon. We followed the [http://partsregistry.org/Help:Standardization| normal process] for converting it to a biobrick. The only complicating factor should have been the large size of the part (11Kbp approx). To help with this we used the stratagene XL Quickchange kit for site dirrected mutagenesis, and XL-10 gold ultra competent cells. In the first mutation test all primer pairs were tested to ensure they functioned, this saved us a week since only a handfull worked without changing the PCR conditions. The gvpQ,gvpA,gvpP were found to be unnecessary by Maura Cannon, but we cleaned them up in parrellel using the pNL26 plasmid with the intension of putting them in annother biobrick.(44A) shows a good sequence.
PCR based insertion of Biobrick prefix and suffix, digestion and ligation to BBa_J61035
Things were going swimmingly until we tried to digest the biobrick prefix & suffix and ligate it into BBa_J61035 which carries a ribosome binding site and gentamycin handle. The digestion, purification, and ligation processes present a large number of options but very few points of intermediate observation. Eventually we obtained a successfull ligation. We think a number of factors were working against the sucessfull ligation. Most significantly digestion of the PstI on the suffix, which is close to the end of the suffix and was surrounded by GC pairs. The small print of the NEB data suggest cutting is inhibitted when the site is surrounded by GC pairs.
The surrounding GC pairs come from the GC clamp we put on the end of the suffix, and the NotI site in the middle of the suffix. This point is worth noting on the suffix design page with a suggestion that AT pairs should be added between the suffix and GC clamp.Due to purchasing of a large number of primers that we didn't want to waste we persisted, swapping to cutting with the more central XbaI and SpeI and accepting the 50% drop in efficiency due to orientation uncertainty. The vector was cut with SpeI and treated with antarctic phosphatase. Details Here
Testing Floatation
- Details:
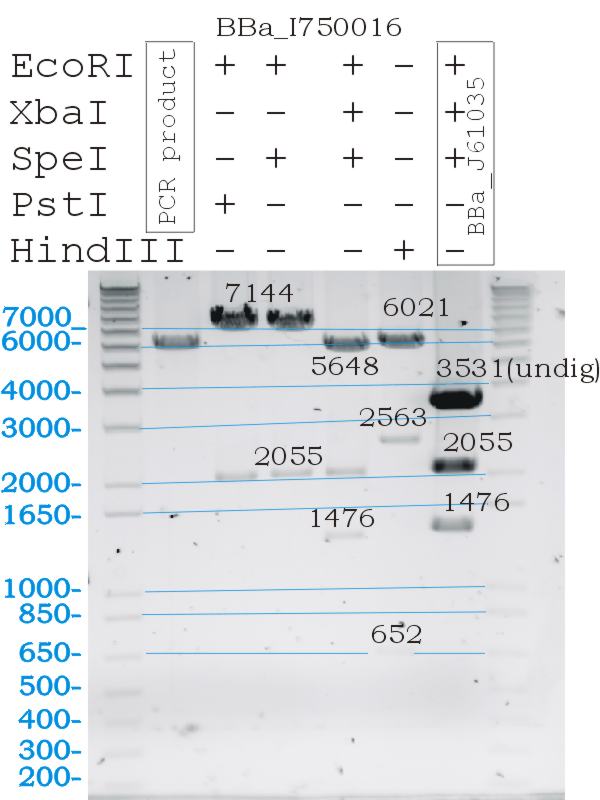
Testing Biobrick restriction sites
- Digests confirmed the correct part size, and biobrick functionallity.
Sequencing
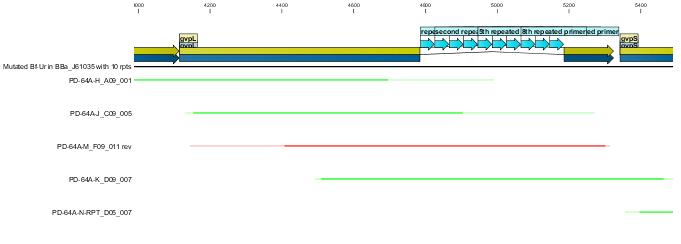
Following the successfull ligation the part was sequenced. In the process we discovered that the 43bp primer used for mutation gvpL(g696a) had self annealed in some way to induce ten repeats of a 40bp sequence.(31A). Interestingly although this introduces a frame shift about 30 amino acids short of the end of GvpL, it does not seem to effect the bouyancy phenotype. The part was documented and shipped as a functional biobrick.
Throughout the process independent parrellel mutation sequences were performed to ensure not single random event could stop the development. (43A) has no g696a induced repeat but the ligation has not been performed on these other parts because of time limmitations resulting from the original ligation failure. There are seven other single nucleotide differences between the sequence and the NCBI reference, but they all turn out to be silent.
Future directions
We produced prefix and suffix primers for each Gvp open reading frame. Once ligation is reliable we intend to produce a sequence of part pairs that when ligated together allow each gene to be knocked out individually. This will we hope lead to a smaller functional biobrick.
The primers are for the coding sequences allowing the RBS to be varied to change the relative expression of each part. With the parts shown to be necessary, it would be possible to use electron microscopy to observe the effects of changing the ammount protein. There is some evidence in the literature to suggest gvpN might be a negative regulator of gas vesicle expression, so knocking this out may generate truely bouyant bacteria, rather than just slow sinkers as is currently observed.
Other work has shown a minimum set of 8 gvp genes is required for gas vesicle expression in annother species, but interestingly these include gvpC and gvpA the closest homolgy genes of which have already be shown not to be necessary in this cluster. Infact gvpL is also closely related to the core protein gvpA so it is quite surprising the frame shift did not disrupt the phenotype. So whats going on?