Paris/August 3
From 2007.igem.org
(Difference between revisions)
(→DGAT biobrick: coR1-Pst1 digestion) |
(→DGAT biobrick: coR1-Pst1 digestion) |
||
| Line 1: | Line 1: | ||
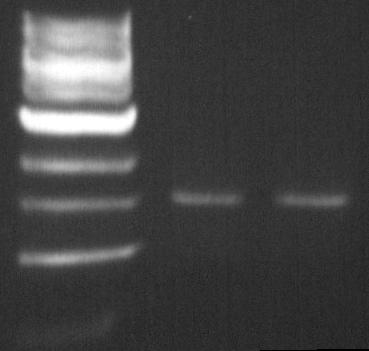
== DGAT biobrick: coR1-Pst1 digestion == | == DGAT biobrick: coR1-Pst1 digestion == | ||
| - | |||
| - | |||
The double digestion allows us to insert the biobrick into biobrick plasmid.<br> | The double digestion allows us to insert the biobrick into biobrick plasmid.<br> | ||
| Line 13: | Line 11: | ||
- Pst-1 enzyme: 1µL<br> | - Pst-1 enzyme: 1µL<br> | ||
- ddH<sub>2</sub>O<br> | - ddH<sub>2</sub>O<br> | ||
| + | |||
| + | [[Image: DGATbbdigEcoR1Pst108032007.jpg|100px|right]] | ||
* '''Protocol:''' | * '''Protocol:''' | ||
Revision as of 10:57, 6 August 2007
DGAT biobrick: coR1-Pst1 digestion
The double digestion allows us to insert the biobrick into biobrick plasmid.
The Pst1 digestion allows us to see if the mutagenesis worked (dgat gene not digested).
- Material:
- DNA (PCR product DGAT deltaPst-1 biobrick from August 2): 200ng = 20µL
- BSA: 0.5µL
- NEBuffer EcoR1: 5µL
- EcoR1 enzyme: 1µL
- Pst-1 enzyme: 1µL
- ddH2O
- Protocol:
- 2 hours at 37°C
- Purification
Quantity:
TOPO cloning
Cloning using TOPO protocol of the following purification of PCR products
| Number | Name | Oligo F | Oligo R | Matrix | Length | Comments |
| P6 | Lox66-DapASubtilis | O8 Lox66-DapASubt-F | O9 DapASubt-R | B.Subtilis 168 | 979 | |
| P7 | Lox71-FtsZ | O1 Lox71-FtsZ-F | O2 FtsZ-R | MG1655 | 1260 | EcoRI site in FtsZ not mutated |
| P8 | RBS-DapAColi | O20 RBS-DapAColi-F | O7 DapAColi-R | MG1655 | 952 | B0030-DapAColi |