Paris/ConstructionProcess
From 2007.igem.org
(Difference between revisions)
Nicolas C. (Talk | contribs) |
Eismoustique (Talk | contribs) (→Overview) |
||
| Line 2: | Line 2: | ||
<br> | <br> | ||
== Overview == | == Overview == | ||
| - | + | ||
| + | |||
| + | Through classical molecular biology techniques, we aimed to construct the following genomically inserted cassette | ||
[[Image:Paris_FinalConstruct.JPG|400px]] | [[Image:Paris_FinalConstruct.JPG|400px]] | ||
Revision as of 18:46, 23 October 2007
Contents |
Overview
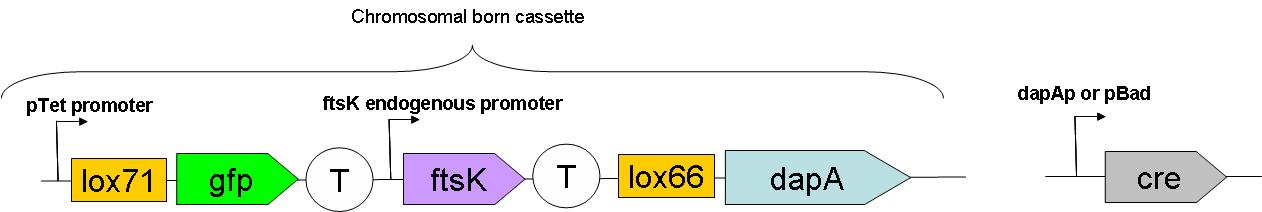
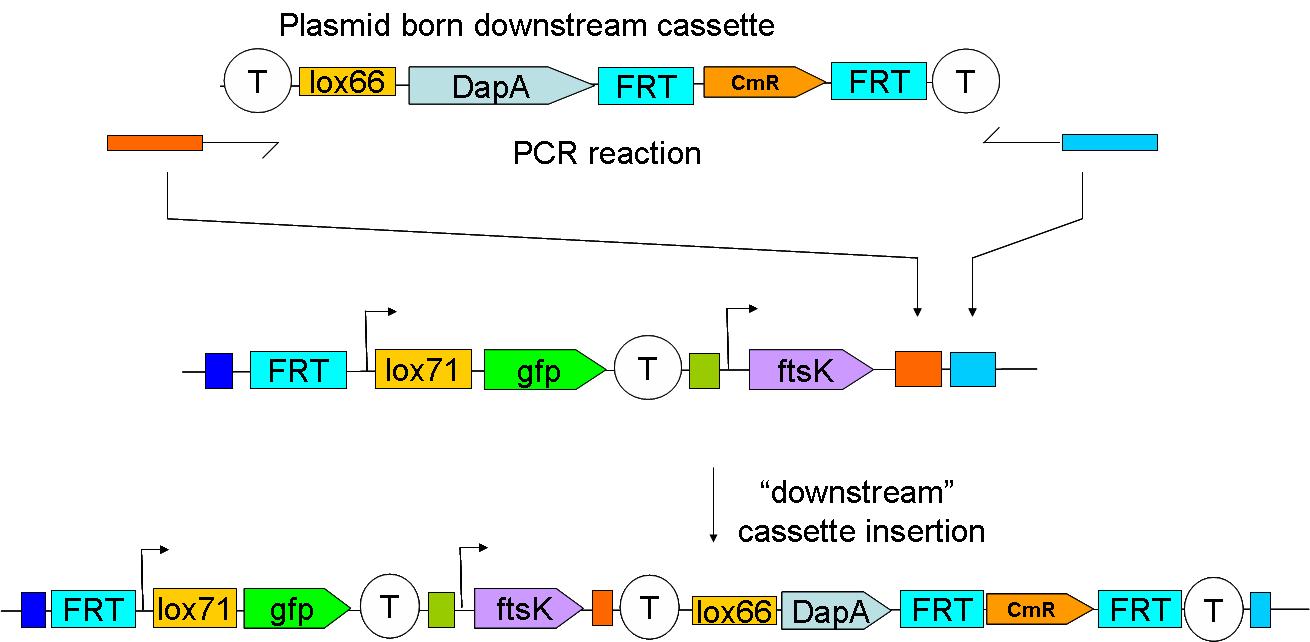
Through classical molecular biology techniques, we aimed to construct the following genomically inserted cassette
In order to construct this genomic cassette, we have considered 2 solutions:
- 1. Either the construct is cloned on a plasmid. Then inserted in the genome strain of a dapA-, ftsK-(ts) E.coli strain. This was the initial strategy we adopted then subsequently abandoned (see here why LIEN).
- 2. Or the cassette is sequentially assembled, in the genome, around the essential gene selected (ftsK in our case).
We have chosen the second strategy. The different steps of this process are as follow:
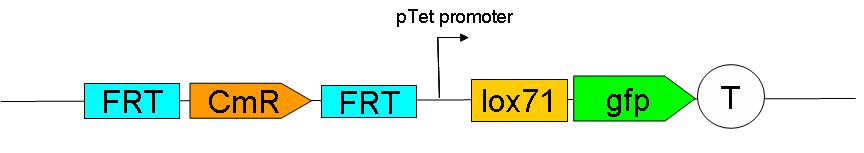
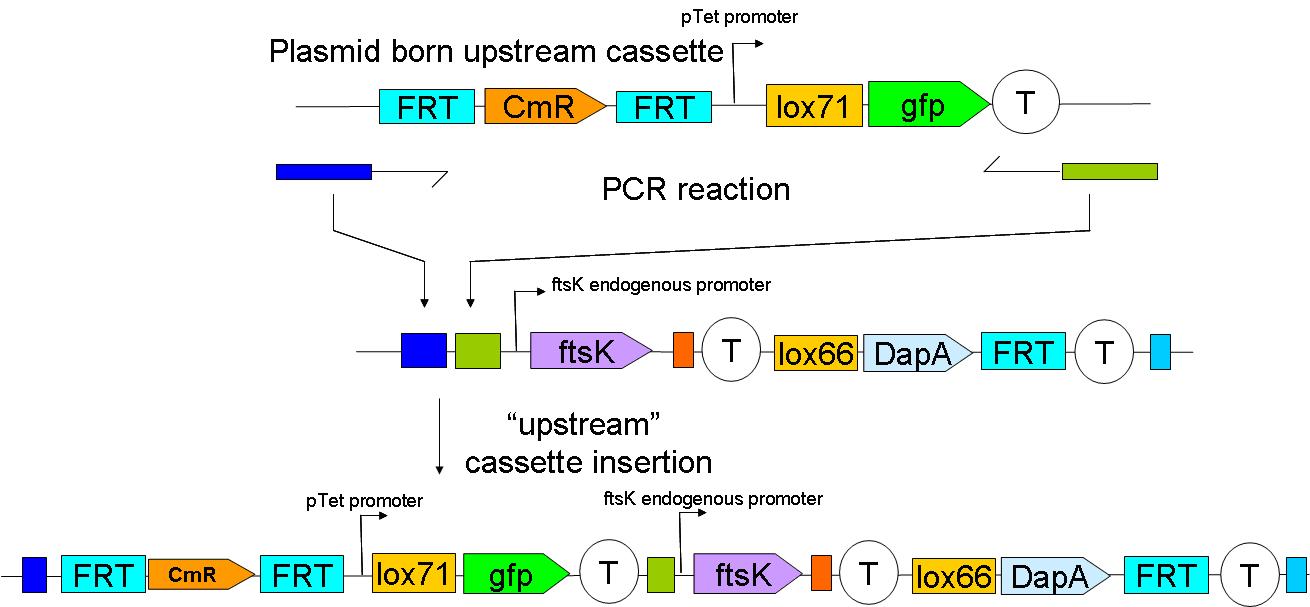
2 plasmid born constructs were constructed. These are termed the UP “upstream construct” & the DP “downstream construct”:
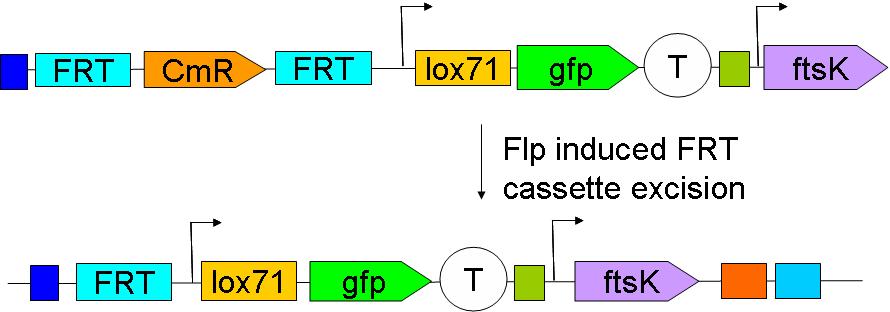
As their names indicate, these constructs are to be inserted upstream and downstream of ftsK in the genome. We chose to insert these constructs using a strategy described by Wanner et al.
We have yet to insert the constructs in the genome. This will be done sequentially:
Constructs
Upstream construct
Downstream construct
Intermediate construct
Backward construct with DapA E.Coli
Backward construct with DapA Subtilis
Recombinaison rate measurement