Paris/ConstructionProcess
From 2007.igem.org
Eismoustique (Talk | contribs) (→Overview) |
Eismoustique (Talk | contribs) (→Overview) |
||
| Line 6: | Line 6: | ||
Through classical molecular biology techniques, we aimed to construct the following genomically inserted cassette | Through classical molecular biology techniques, we aimed to construct the following genomically inserted cassette | ||
| - | [[Image:Paris_FinalConstruct.JPG| | + | [[Image:Paris_FinalConstruct.JPG|500px]] |
This full construct has yet to be assembled in the genome. | This full construct has yet to be assembled in the genome. | ||
| Line 20: | Line 20: | ||
constructed. These are termed the “'''upstream construct'''” & the “'''downstream construct'''”: | constructed. These are termed the “'''upstream construct'''” & the “'''downstream construct'''”: | ||
| - | [[Image:Paris_Recombination scheme.JPG| | + | [[Image:Paris_Recombination scheme.JPG|600px]] |
| Line 53: | Line 53: | ||
'''Upstream construct:''' | '''Upstream construct:''' | ||
| - | [[Image:Paris_UpstreamConstruct.JPG| | + | [[Image:Paris_UpstreamConstruct.JPG|500px]] |
See here (LIEN vers une page avec la construction de Nico), how this was sequentially assembled on a plasmid. | See here (LIEN vers une page avec la construction de Nico), how this was sequentially assembled on a plasmid. | ||
'''Downstream construct:''' | '''Downstream construct:''' | ||
| - | [[Image:Paris_DownstreamConstruct.JPG| | + | [[Image:Paris_DownstreamConstruct.JPG|500px]] |
See here (LIEN vers une page avec la construction de Nico), how this was sequentially assembled | See here (LIEN vers une page avec la construction de Nico), how this was sequentially assembled | ||
| Line 73: | Line 73: | ||
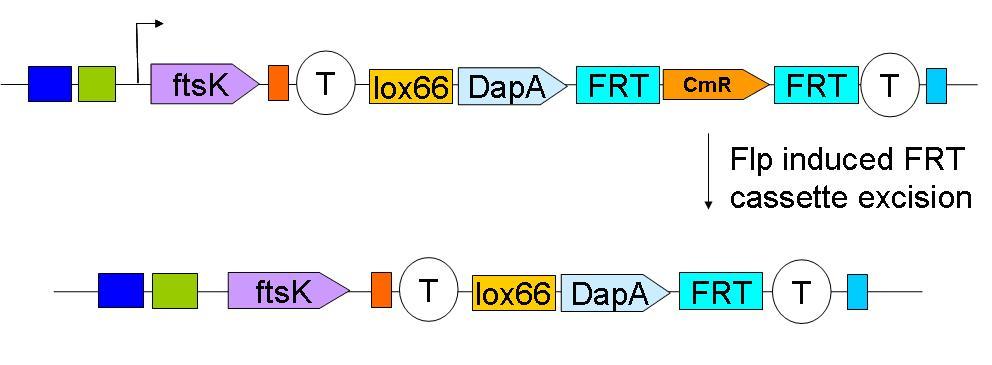
In a first step the downstream construct is amplified using oligonucteotides baring sequences homologous to those downstream of ftsK. The PCR product is used to transform a dapA- E.coli strain. | In a first step the downstream construct is amplified using oligonucteotides baring sequences homologous to those downstream of ftsK. The PCR product is used to transform a dapA- E.coli strain. | ||
| - | [[Image:Paris_Downstream Insertion.JPG| | + | [[Image:Paris_Downstream Insertion.JPG|500px]] |
| Line 83: | Line 83: | ||
| - | [[Image:Paris_Flp.JPG| | + | [[Image:Paris_Flp.JPG|500px]] |
| Line 89: | Line 89: | ||
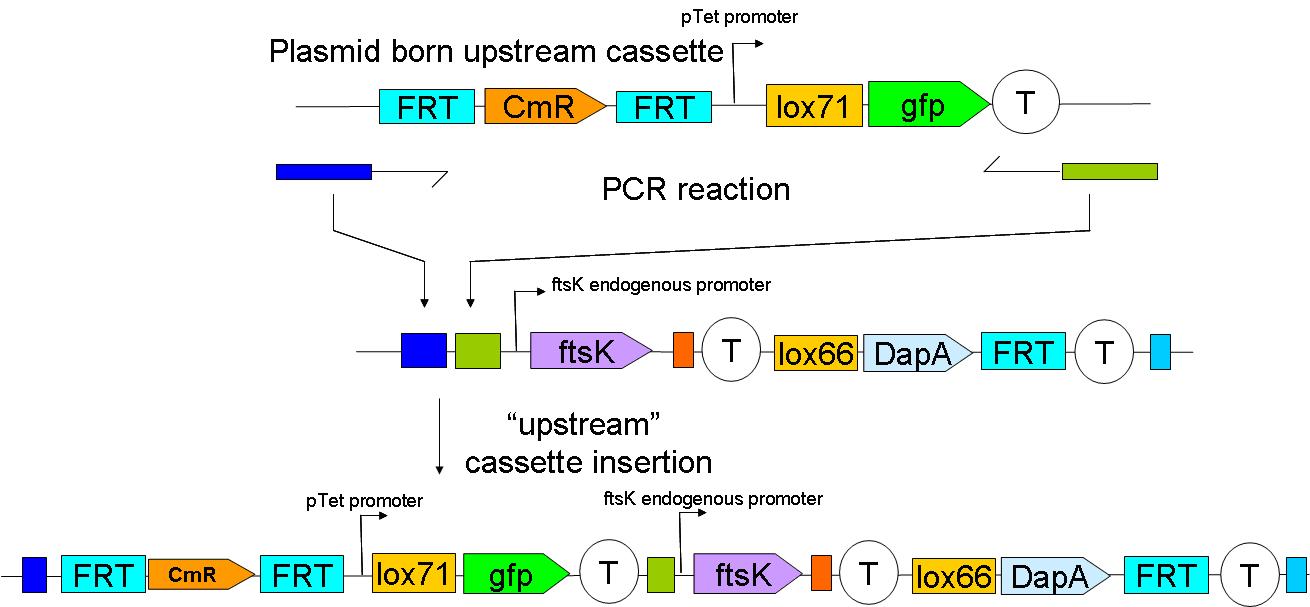
| - | [[Image:Paris_UpstreamInsertion.JPG| | + | [[Image:Paris_UpstreamInsertion.JPG|700px]] |
== Constructs == | == Constructs == | ||
Revision as of 21:12, 23 October 2007
Contents |
Overview
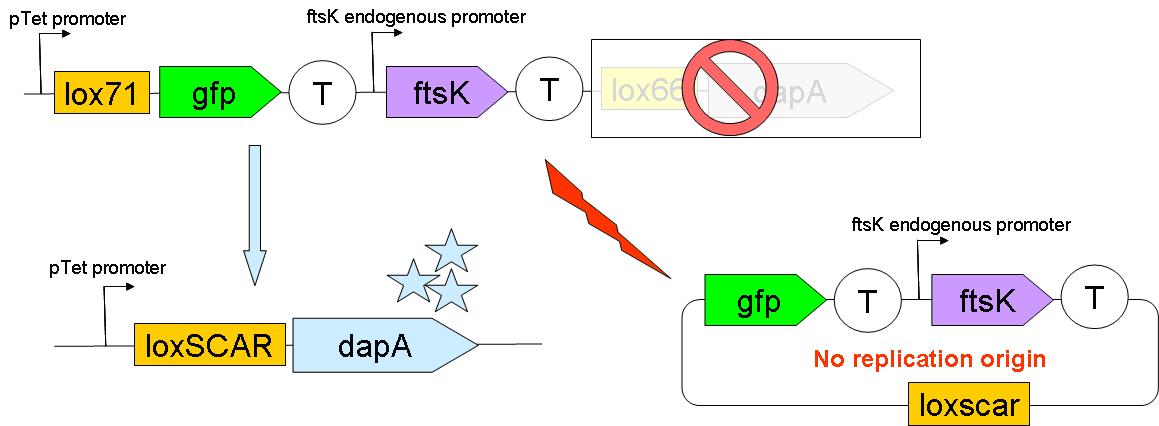
Through classical molecular biology techniques, we aimed to construct the following genomically inserted cassette
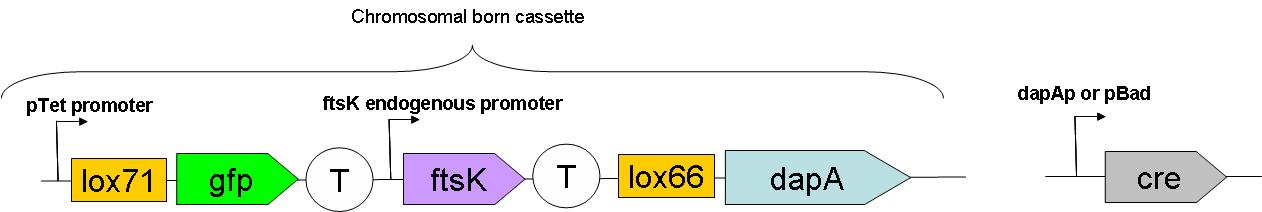
As can be seen on this schematic representation of the SMB genomic backbone cassette, at basal genomic state (in Germ line cells):
- pTet promoter drives the expression of gfp.
- The expression of ftsK is controlled by its natural promoter.
- ftsK is isolated from pTet promoter by the intercalation of Terminator (B0015 terminator).
- dapA gene is in a dormant state since it lacks a promoter to drive its transcription.
Upon G to S differentiation, the following genomic reassembly should take place:
constructed. These are termed the “upstream construct” & the “downstream construct”:
Cre mediated lox recombination should lead to:
- excision of ftsK gene from the genome on a circular unreplicative DNA molecule. We ignore how much time such a DNA is stable in a cell.
- placing dapA gene under the control of pTet promoter. This should lead to dapA expression, & hopefully to DAP synthesis.
In order to construct this genomic cassette, we have considered two technical solutions:
1) Either the full construct is cloned on a plasmid and inserted in the genome of a dapA-, ftsK-(ts) E.coli strain. This was the initial strategy we adopted then subsequently abandoned (see here why LIEN).
2) Or the cassette is sequentially assembled, in the genome, around the essential gene selected (ftsK in our case).
The second strategy does not require cloning ftsK gene & does not require using an ftsK mutant.
We have chosen the second strategy mentioned above to generate the SMB genomic backbone construct. The different steps of this process are as follow:
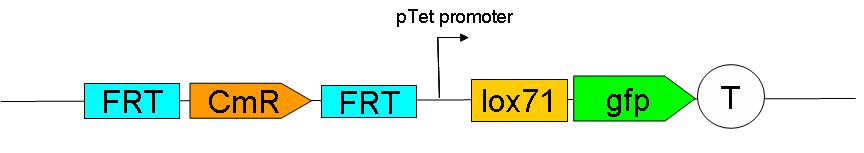
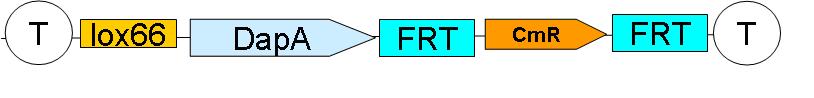
2 plasmid born constructs were generated. These are termed the “upstream construct” & the “downstream construct”:
Upstream construct:
See here (LIEN vers une page avec la construction de Nico), how this was sequentially assembled on a plasmid.
See here (LIEN vers une page avec la construction de Nico), how this was sequentially assembled
As their names indicate, these constructs are to be inserted upstream and downstream of ftsK gene in the genome of a dapA- strain.
We chose to insert these constructs using a strategy described by …. et al. (ref Wanner)
Petite description de la stratégie de Wanner a mettre a faire.
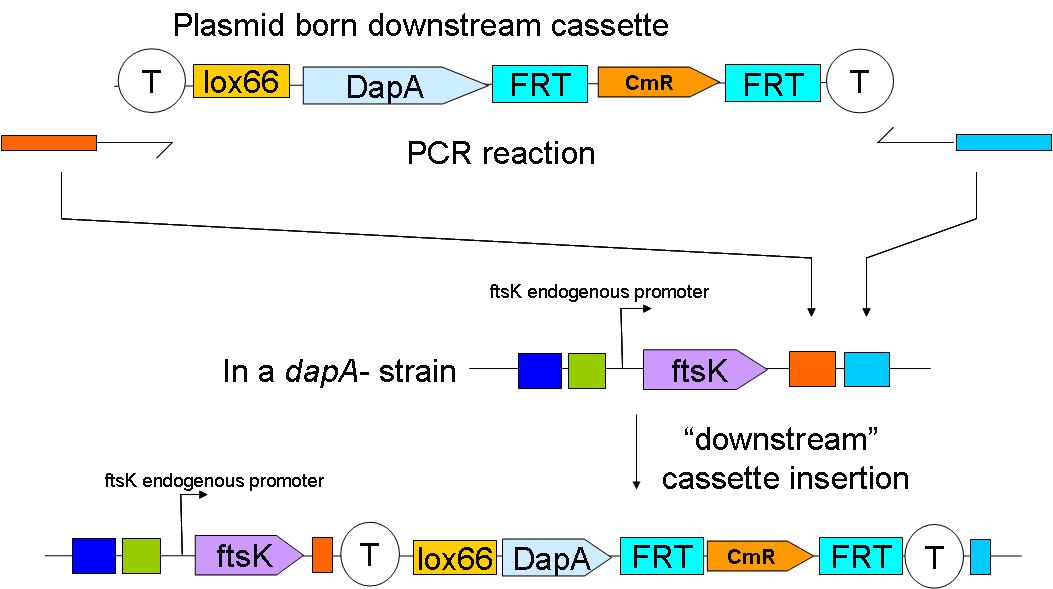
We have yet to insert the constructs in the genome. This will be done sequentially:
In a first step the downstream construct is amplified using oligonucteotides baring sequences homologous to those downstream of ftsK. The PCR product is used to transform a dapA- E.coli strain.
The resistance cassette frt-CmR-frt carried by the downstream construct allows screening for the event of genomic insertion of the construct. CmR gene confers resistance to the antibiotic chloramphenicol.
Subsequently, the Chloramphenicol resistance gene is eliminated by FRT recombination. This reaction is to be catalysed by the site specific recombinase: Flp.
The upstream construct can then be inserted based on the same strategy.
Constructs
Upstream construct
Downstream construct
Intermediate construct
Backward construct with DapA E.Coli
Backward construct with DapA Subtilis
Recombinaison rate measurement