Paris/Continuous modelb
From 2007.igem.org
(→Robustness) |
|||
| Line 89: | Line 89: | ||
<br> | <br> | ||
| + | |||
| + | |||
== Analysis == | == Analysis == | ||
| Line 94: | Line 96: | ||
In order to evaluate the robustness of both models, we try to find a box in the parameter space containing p* such that for any poin of this box the specification we defined is valid. | In order to evaluate the robustness of both models, we try to find a box in the parameter space containing p* such that for any poin of this box the specification we defined is valid. | ||
| + | |||
| + | [[Image:Robref.jpg|left|450px|refGS]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
=== Optimization === | === Optimization === | ||
Revision as of 14:32, 24 October 2007
We developed simple models to evaluate the relative benefits of two different designs in term of robustness and optimization capabilities.
Contents |
Introduction
This model aims at describing the dynamic evolution of populations of germen and soma type bacteria. It is based on a set of differential equations describing DAP synthesis, DAP transport, differentiation of germen bacteria into soma and bacteria death. This approach differs form the precedents one by the level of description of the model and the numerical analysis done on the model.
By conducting robustness and optimization analysis on two different systems, one with a constant rate of differentiation against one with a rate of differentiation driven by the concentration of DAP, we evaluate benefits and drawbacks of both systems.
We first display the consistency of the biological system by providing a set of kinetic parameters such that the numerical simulation validate a given minimal behavior. Then we analyze the system robustness with regard to its kinetic parameters and finally we try to optimize the system output by adjusting some biologically relevant parameters.
Problem description
System
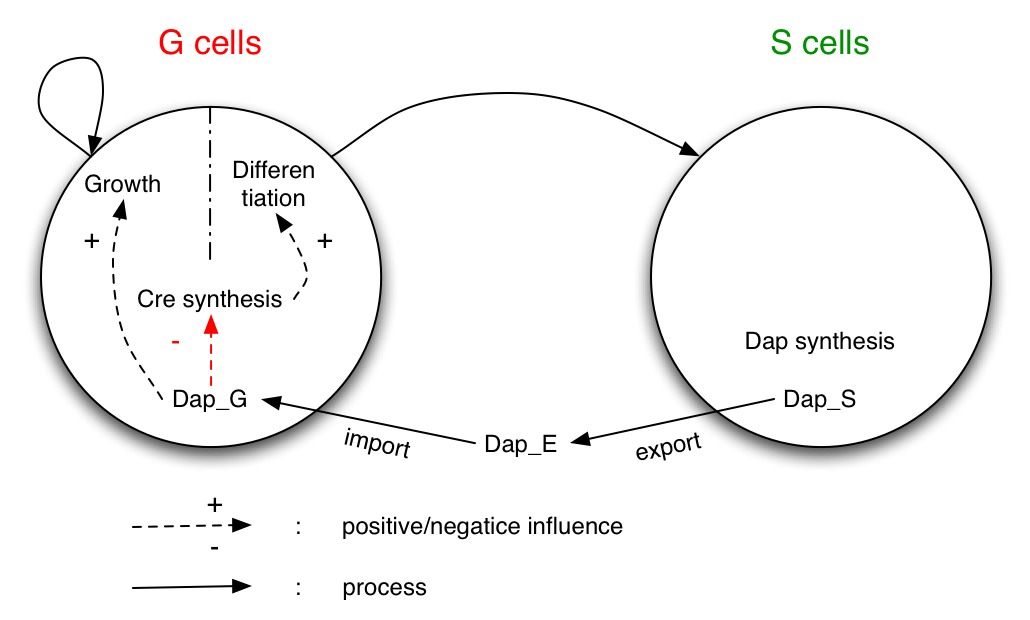
The system studied is made of two populations of bacteria G and S. Bacteria G can either divide or differentiate into S.
Dap is only synthesized by bacteria S and diffuses in the environment and in G.
G is dependent of Dap for division. Differentiation of G is controlled by Cre. Cre synthesis in G is either constant or dependant of Dap (red arrow).
Expected behavior
We consider initial conditions in which there is some bacteria G and no soma type bacteria, so that the system first has to go through an intialisation phase where a part of the initial population of G differentiate into S. Adding Dap in the environment can only have a benefic effect on the population so we choose to start this study with an initial condition lacking Dap. The requested behavior for this system is that it grows fast enough and has a robust initialization phase.
Modeling purpose
- Is minimal behavior robust ?
- How can we tune the system to improve its output ?
- Which system should we prefer ?
Modelling
Differential equations
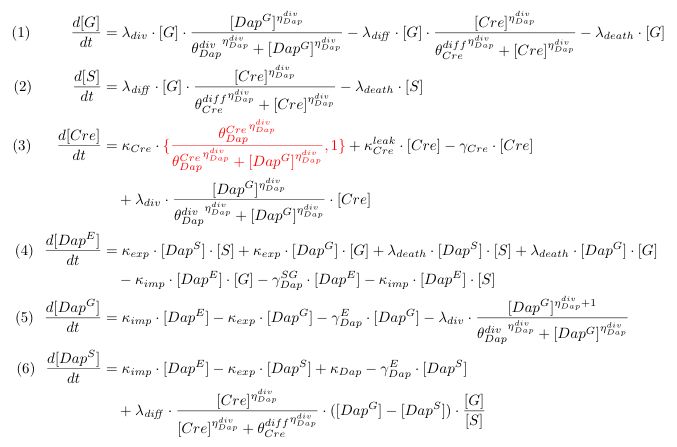
To model the dependence of division and differentiation of G we use Michaelis-Menten kinetics (terms λdiv and λdiff in equations 1 and 2). Equation (3) represents the synthesis of Cre and its degradation. We consider two models : the synthesis rate of Cre is either constant or inhibited by DAP (red term).
Term λdiv in equations (3,4) accounts for the dilution of Cre and DAP when the population of G grows.Equations involving DAP (4,5,6) represent DAP synthesis in somabacteria, DAP degradation in bacteria and in the environment, DAP transport and the release of DAP in the environment from dying bacteria. Terms λdiff in equations (4,5) are corrective terms to take into account the mean variation of concentration of DAP in G and S when some bacteria G differentiate into S.
Parameters
We first try to find out a set of paramater values such that the system exhibits a minimal growth and a robust initialization phase. We will then use this set of parameter values as a reference point for the robustness and optimization analysis.
Initial condition
The initial condition used as reference is made of some bacteria G, no bacteria S, no Dap and no Cre.
Kinetic Parameters
In order to set parameter values we use three different methods. First, parameters λdiv and λdeath are estimations based on experimental data. Then, we fix the following inequality constraints : κimp > κexp, λdeath<λdiv, λdiff > λdiv and γDapE < γDapGS. These inequalities express biological knowledge : Dap is actively imported into bacteria, bacteria death is lower than bacteria growth, differentiation must be lower than bacteria growth and degradation of DAP in the environment is lower than in bacteria where it is consumed. Finally we impose order of magnitude constraints to express reaction speed differences : κexp,κimp>> λdiv,λdeath,λdiff means that transport of DAP is an order of magnitude faster than reactions modelling death, growth or differentiation of bacteria.
Based on these constraints on the parameters, we defined a set of parameter values p* as a reference starting point.
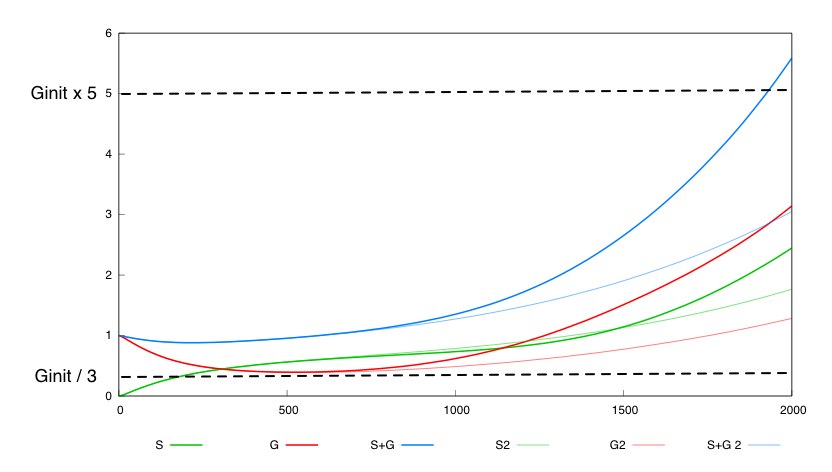
Specification
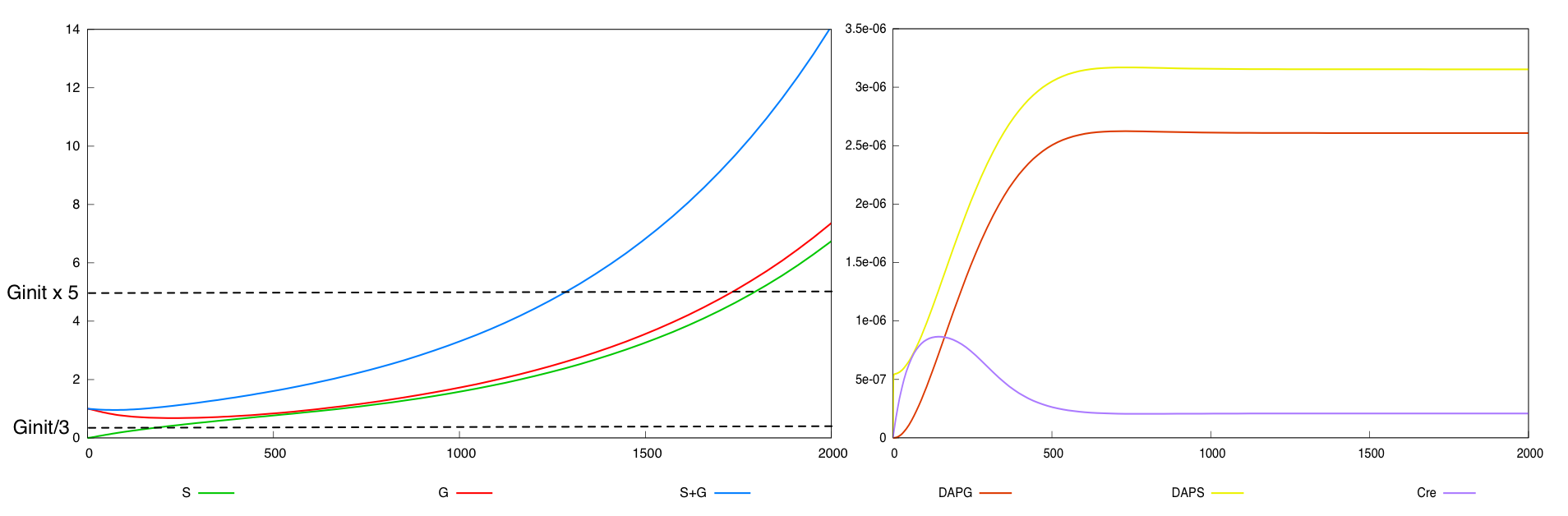
We define a minimal behavior that the system must display. First, we want that the quantity of bacteria G never
falls below threshold Ginit/3. Secondly, we want that after a certain amount of time (2000 time units), the overall population of bacteria (G+S) is above threshold Ginit*5.
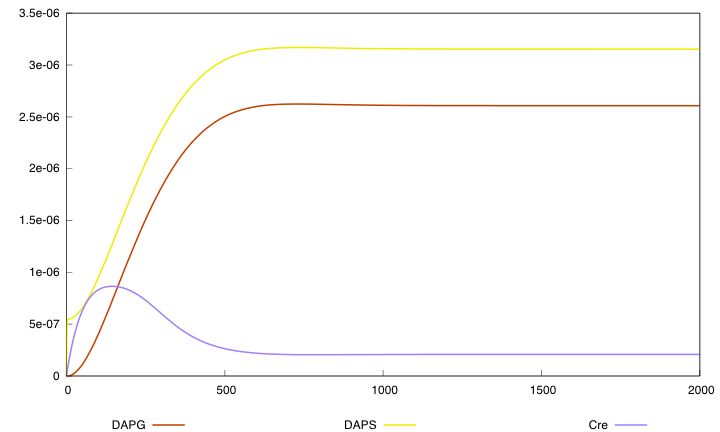
For the parmaeter values p*, both systems validate this specification. The system with a synthesis rate of Cre dependent of Dap has the following behavior :
Analysis
Robustness
In order to evaluate the robustness of both models, we try to find a box in the parameter space containing p* such that for any poin of this box the specification we defined is valid.