Paris/July 13
From 2007.igem.org
(→Transduction of MG1655 with P1 stock made on w121) |
Nicolas C. (Talk | contribs) |
||
| (25 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Paris/July 12|yesterday]] -- [[Paris/July 14|tomorrow]] <br> | ||
| + | == Test of Ftsz TS strain == | ||
| + | The isolated clones of FtsZ TS all grew at 42°C and at 30°C. They must be mutants... So next time we'll try to find other clones from the source of the strain. | ||
== Transduction of MG1655 with P1 stock made on w121 == | == Transduction of MG1655 with P1 stock made on w121 == | ||
| - | + | We used 03/07/07 (I) and 12/07/07 (II) phage stock from W121 and MG1655 culture overnight. | |
| + | - Control (1mL LB MgSO4 30mM; CaCl2 15mM) <br> | ||
| + | - 5µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight <br> | ||
| + | - 50µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight <br> | ||
| + | - 500µL Phage + 500µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight <br> | ||
| + | |||
| + | See [[Paris/PROTOCOLS#Transduction_to_MG1655_using_the_P1_stock_made_on_w121.|Protocols]] | ||
| + | |||
| + | == Transformation of Biobricks from the iGEM2007 plates into DH5aplha == | ||
| + | * <bbpart>BBa_I0500</bbpart>: Inducible pBad/araC in pSB2K3 (KanR) | ||
| + | well: 9I, Plate 2 | ||
| + | |||
| + | * <bbpart>BBa_J23100</bbpart>: strong constitutive promoter in BBa_J61002 (AmpR) | ||
| + | well: 21E, Plate 3 | ||
| + | |||
| + | * <bbpart>BBa_B0015</bbpart>: double terminator (B0010-B0012) in pSB1AK3 (AmpR) | ||
| + | well: 1I, Plate 1 | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | + | Unfreeze chemio-competent bacteria (E.coli DH5alpha; Invitrogen) on ice 5 min<br> | |
| - | + | Add 1µL of each resuspended plasmid of interest into 50µL of precedent competent bacteria on ice<br> | |
| - | + | Incubate on ice 30 min<br> | |
| - | + | heat Shock cells for 20 sec in a 42°C water bath without shaking<br> | |
| - | + | Place tube on ice for 2 min<br> | |
| + | Add 950µL of pre-waemed medium of choice to each tube<br> | ||
| + | Incubate tubes at 37°C for 1 hour at 200 rpm<br> | ||
<br> | <br> | ||
| - | + | Spread 20 and 200µL from each transformation on pre-warmed selective plates.<br> | |
| + | * <bbpart>BBa_I0500</bbpart>: Inducible pBad/araC on KanR (100µg/mL) LB-agar | ||
| + | * <bbpart>BBa_J23100</bbpart>: strong constitutive promoter on AmpR (100µg/mL) LB-agar | ||
| + | * <bbpart>BBa_B0015</bbpart>: double terminator (B0010-B0012) on AmpR(100µg/mL) LB-agar | ||
| + | <br> | ||
| + | Incubate plates ON at 37°C<br> | ||
| + | <br> | ||
| + | |||
| + | == Microscopy of Acinobacter strain == | ||
| + | We learn how to use the microscope to see single cells (see [[Paris/PROTOCOLS#Fluorescent_single_cells_visualisation| Protocols]]). | ||
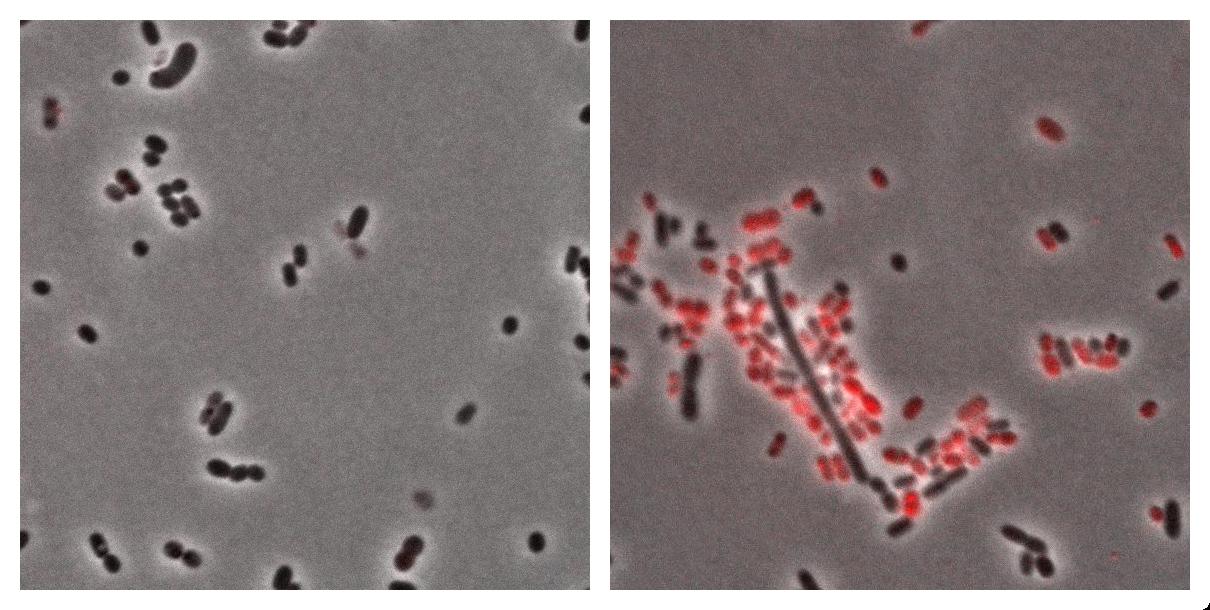
| + | Here we show the photos taken from Acinobacter cells <br> | ||
| + | * left : Acinetobacter grown in Low Nitrogen Mineral medium. | ||
| + | * right : Acinetobacter grow in Low Nitrogen Mineral medium with Nile Red . | ||
| + | Exposure time and scaling is the same for both images. Clearly, some cells on the right are stained by Nile Red, which is specific to triglycerides, whereas almost all cells on the left are not stained. Tipically, the size of one cell is about 5µm. | ||
| + | [[Image:Paris_Acineto_NileRed.jpg|850px]] | ||
| + | <br> | ||
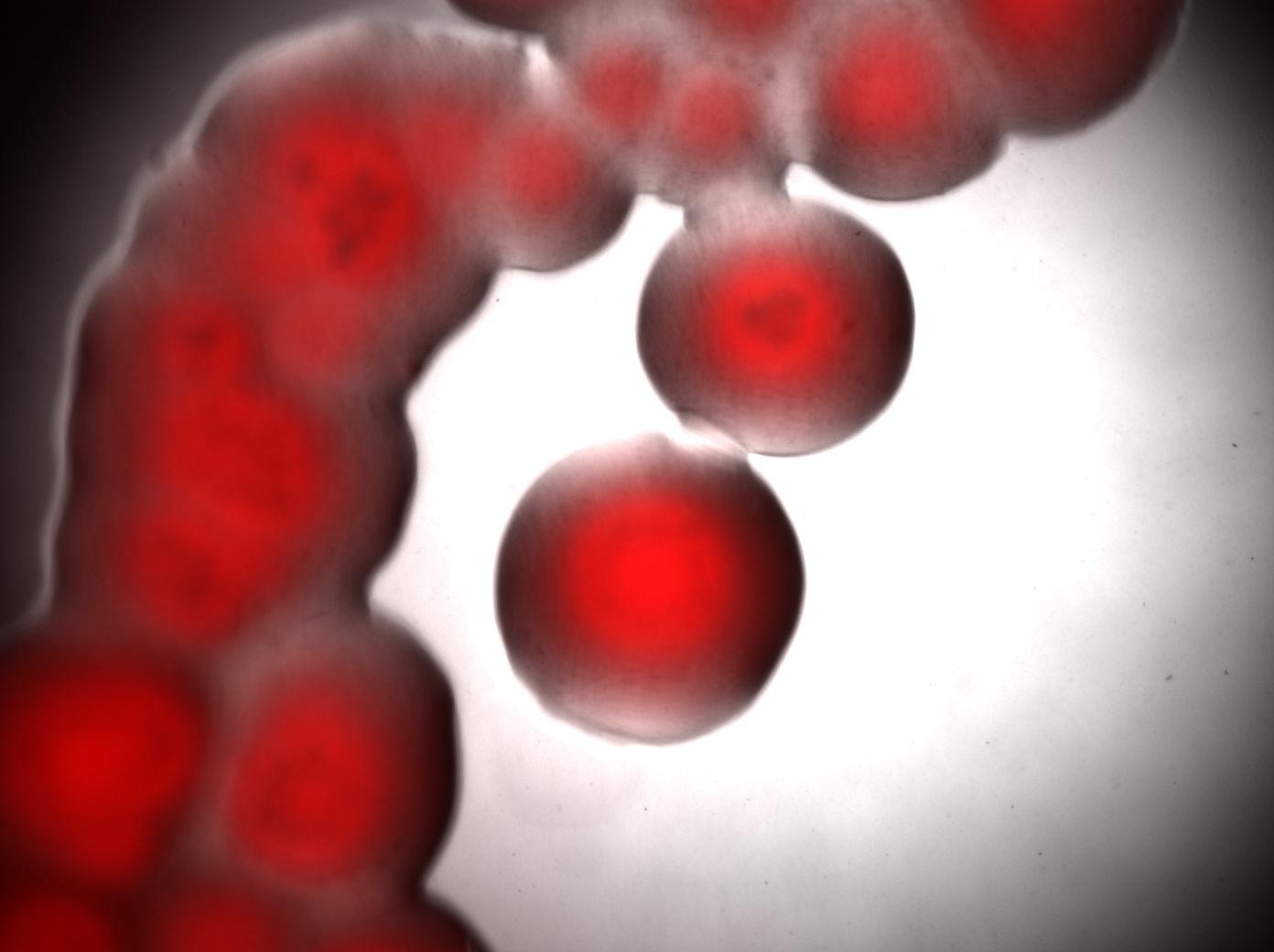
| + | Bottom image : visualisation of colonies stained by Nile Red. It seems that only cells in the middle of colonies (i.e. "old" cells) are producing triglycerides whereas cells on the edges doesn't. <br> | ||
| + | [[Image:Paris_Acineto_Colonies.jpg|500px|center]] | ||
| + | |||
| + | == PCRs == | ||
| + | We received the oligos !!! But not all of them :-( | ||
| + | |||
| + | 3 PCRs: | ||
| + | * Lox71-FtsA-FtsZ1 : 3 (Lox71-FtsA-F) + 4 (DEcoR1-FtsZ-R) | ||
| + | * FtsZ2: 5 (DEcoR1-FtsZ-F) + 2 FtsZ-R | ||
| + | * Lox66-DapAColi: 6 (Lox66-DapAColi-F) + 7 (DapAColi-R) | ||
| + | |||
| + | ==Titration of bacteriophages P1 == | ||
| + | We measure the strength of former P1 preparation : | ||
| + | * 3.7.7 : ~10<sup>5</sup>/ml : very bad | ||
| + | * 8.7.7 : ~10<sup>2</sup>/ml : very bad | ||
| + | * 12.7.7 : pb : we had the phages too late, and we can't conclude. | ||
Latest revision as of 17:46, 7 October 2007
Contents |
Test of Ftsz TS strain
The isolated clones of FtsZ TS all grew at 42°C and at 30°C. They must be mutants... So next time we'll try to find other clones from the source of the strain.
Transduction of MG1655 with P1 stock made on w121
We used 03/07/07 (I) and 12/07/07 (II) phage stock from W121 and MG1655 culture overnight.
- Control (1mL LB MgSO4 30mM; CaCl2 15mM)
- 5µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight
- 50µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight
- 500µL Phage + 500µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture overnight
See Protocols
Transformation of Biobricks from the iGEM2007 plates into DH5aplha
- BBa_I0500: Inducible pBad/araC in pSB2K3 (KanR)
well: 9I, Plate 2
- BBa_J23100: strong constitutive promoter in BBa_J61002 (AmpR)
well: 21E, Plate 3
- BBa_B0015: double terminator (B0010-B0012) in pSB1AK3 (AmpR)
well: 1I, Plate 1
Unfreeze chemio-competent bacteria (E.coli DH5alpha; Invitrogen) on ice 5 min
Add 1µL of each resuspended plasmid of interest into 50µL of precedent competent bacteria on ice
Incubate on ice 30 min
heat Shock cells for 20 sec in a 42°C water bath without shaking
Place tube on ice for 2 min
Add 950µL of pre-waemed medium of choice to each tube
Incubate tubes at 37°C for 1 hour at 200 rpm
Spread 20 and 200µL from each transformation on pre-warmed selective plates.
- BBa_I0500: Inducible pBad/araC on KanR (100µg/mL) LB-agar
- BBa_J23100: strong constitutive promoter on AmpR (100µg/mL) LB-agar
- BBa_B0015: double terminator (B0010-B0012) on AmpR(100µg/mL) LB-agar
Incubate plates ON at 37°C
Microscopy of Acinobacter strain
We learn how to use the microscope to see single cells (see Protocols).
Here we show the photos taken from Acinobacter cells
- left : Acinetobacter grown in Low Nitrogen Mineral medium.
- right : Acinetobacter grow in Low Nitrogen Mineral medium with Nile Red .
Exposure time and scaling is the same for both images. Clearly, some cells on the right are stained by Nile Red, which is specific to triglycerides, whereas almost all cells on the left are not stained. Tipically, the size of one cell is about 5µm.
Bottom image : visualisation of colonies stained by Nile Red. It seems that only cells in the middle of colonies (i.e. "old" cells) are producing triglycerides whereas cells on the edges doesn't.
PCRs
We received the oligos !!! But not all of them :-(
3 PCRs:
- Lox71-FtsA-FtsZ1 : 3 (Lox71-FtsA-F) + 4 (DEcoR1-FtsZ-R)
- FtsZ2: 5 (DEcoR1-FtsZ-F) + 2 FtsZ-R
- Lox66-DapAColi: 6 (Lox66-DapAColi-F) + 7 (DapAColi-R)
Titration of bacteriophages P1
We measure the strength of former P1 preparation :
- 3.7.7 : ~105/ml : very bad
- 8.7.7 : ~102/ml : very bad
- 12.7.7 : pb : we had the phages too late, and we can't conclude.