Paris/July 22
From 2007.igem.org
(Difference between revisions)
(→E.Coli pKS::DGAT) |
(→E.Coli pKS::DGAT) |
||
| Line 27: | Line 27: | ||
* Interpretation: | * Interpretation: | ||
| - | We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences. | + | We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences. Signal would be better with more cells. |
Revision as of 15:43, 30 July 2007
Digestion products
Purification of the digestion products of 21/07/07 purif. using the SV wizard gel cleanup system
elution of the purification products in 30-40microL H2O
Overnight cultures launched
The following ON cultures were launched:
- pJ23107 clone 3 (from the glycerol stock S14.3) in 5ml LB-Amp. For performing a miniprep
- w121 (from glycerol stock S12) in 5 ml LB-Erythromycin. For performing growth-kinetics analysis on DAP supplemented media
I was unable to launch the following culture because of lack of a glycerol stock:
- pJ23107 clone 2 (from the glycerol stock S6.2) in 5ml LB-Amp. For performing a miniprep
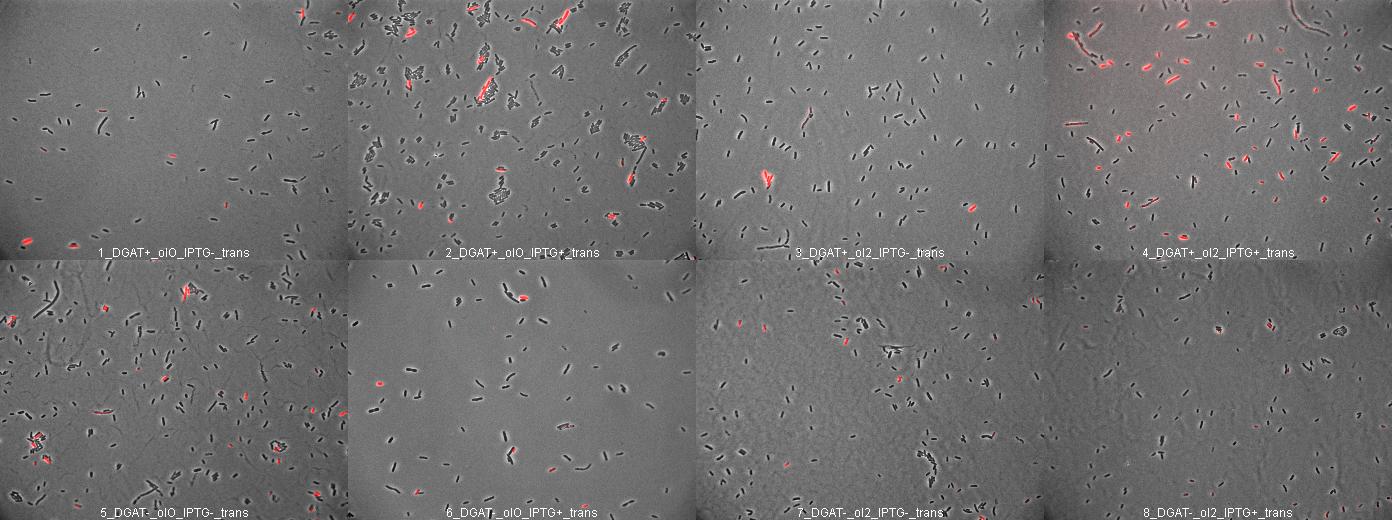
E.Coli pKS::DGAT
We look under microscopy 72 hours after incubation of E.coli transformed by pKS::DGAT and the control E.coli transformed by part B0015 on different LB medium (See July 18).
- Observation:
We can observe E.coli single cells (100X).
- Interpretation:
We can observe lipid inclusion into E.coli transformed by pKS::DGAT with IPTG induction. With or without adding oleate we do not observe significant differences. Signal would be better with more cells.