Tokyo/Expression level check
From 2007.igem.org
(→Result & Conclusion:) |
|||
| Line 25: | Line 25: | ||
<br>Based on the results, the ranges of bistability and those of mono-stability are calculated as shown in Fig. 1. | <br>Based on the results, the ranges of bistability and those of mono-stability are calculated as shown in Fig. 1. | ||
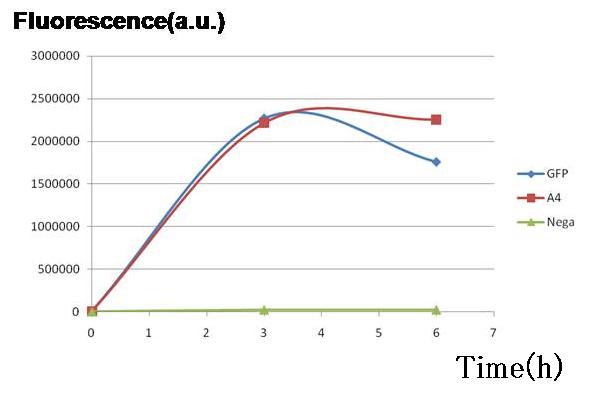
<br>.[[Image:pC1GFP.jpg|thumb|350px| '''Fig.1''' Genes downstream of the promoters on the both sides were expressed to almost the same degrees in terms of GFP fluorescence.|left]] | <br>.[[Image:pC1GFP.jpg|thumb|350px| '''Fig.1''' Genes downstream of the promoters on the both sides were expressed to almost the same degrees in terms of GFP fluorescence.|left]] | ||
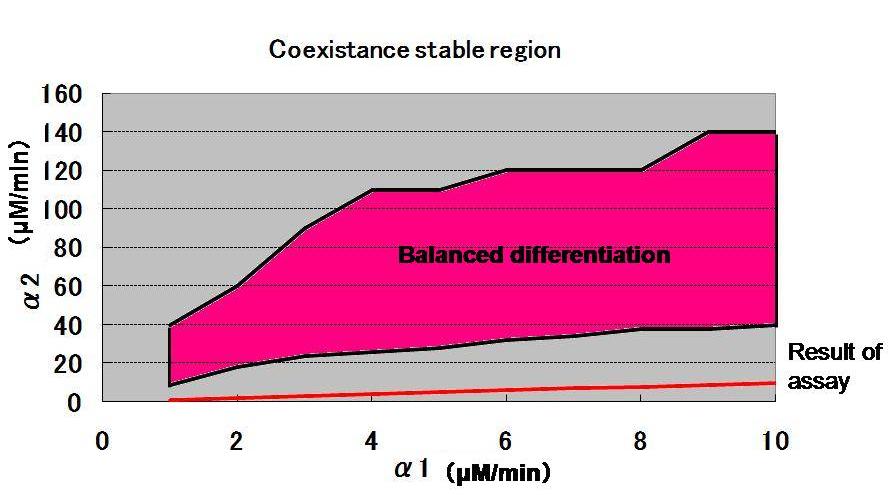
| - | [[Image: | + | [[Image:Balanced.jpg|thumb|500px|'''Fig.2 Calculated coexistance stable region'''|center|left]] |
Revision as of 02:57, 27 October 2007
Works top 0.Hybrid promoter 1.Formulation 2.Assay1 3.Simulation 4.Assay2 5.Future works
Activation check by cell-produced AHL Expression level check on promoters + plasmid sets of A and B sides
Expression level check on two different promoters + plasmid sets
Objective:
To measure and compare the activities of two different promoters + plasmid sets by fluorescence of GFP downstream of each promoter. Lambda cI-regulated promoter and the lux lac hybrid promoter were tested.
Samples:
Each sample cells has two plasmids.
- A4ΔP(BBa_I751100) / pc1-GFP (BBa_I751311)
- A4 hybrid promoter(BBa_I751101) / pBR322TetR (+)AHL
- A4 hybrid promoter(BBa_I751101) / pBR322TetR (-)AHL
*The promoter pcI sequence was designed to contain OR1, -35, and -10 regions, but not R2 or OR3.
Procedure:
AHL assay Standard protocol
Wash
OD and fluorescence were measured 0, 3, and 6 hours after the fresh culture incubation started.
Result & Conclusion:
Two plasmid sets, A4Δp+pc1-GFP and A4 hybrid+GFP PBR322TetR (+)AHL, shows almost the same fluorescence of GFP, indicating that expression levels of both sets are almost the same though the latter is a bit smaller.
Based on the results, the ranges of bistability and those of mono-stability are calculated as shown in Fig. 1.
.