Toronto/Project Description
From 2007.igem.org
ElliottSdA (Talk | contribs) m (→SIMULATION) |
|||
| Line 7: | Line 7: | ||
=== OBJECTIVES === | === OBJECTIVES === | ||
| - | The goal of the proposed project is to develop a simple two- | + | The goal of the proposed project is to develop a simple two-input-one-output feedforward NN using E. coli cells. This network will be capable of being trained to function as different types of digital logic gates. |
=== REVIEW OF PERTINENT LITERATURE === | === REVIEW OF PERTINENT LITERATURE === | ||
| Line 23: | Line 23: | ||
=== METHODS === | === METHODS === | ||
| - | '''''A. Basic E. Coli Neural Network | + | '''''A. Basic E. Coli Neural Network Structure''''' |
| - | + | The input to the proposed network will be light and the output will be ECFP expression. Two red light sources will comprise the input nodes. The cell population will consist of output cells and weight cells. The output cells, which represent the output node, will be physically separated into three neighboring groups by a membrane to prevent cell migration. The first two groups detect light from the two network inputs. The weight cells will be mounted above two groups of output cells and will act as a light filter to control the amount of light received by the output cells. The weight cell groups will be separated via a membrane to prevent cell migration. The amount of light picked up by each individual output cell group is integrated via production of HSL signaling molecules. These molecules are picked up by all output cell groups. The third group of output cells has no weight cells above it and exists only for the measurement of ECFP expression. For each input-output measurement and between training sets, the media should be replaced to remove HSL and blue substrate molecules. An abstracted diagram of the topology is illustrated in Fig. 2. The genetic parts for the output cells are shown in Fig. 3. | |
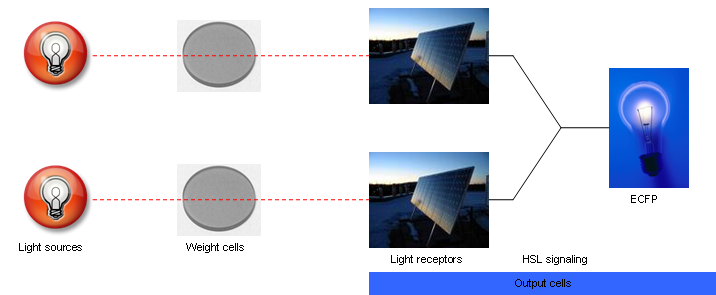
| - | + | [[Image:fig2.png|frame|center|Fig. 2. Abstracted diagram of the proposed neural network structure.]] | |
| - | The | + | [[Image:fig3.png|frame|center|Fig. 3. The genetic parts for constructing the output cells.<br> (a) The light receptor is constitutively expressed.<br> (b) OmpR is activated in darkness when the EnvZ domain on the light receptor is phosphorylated. The activation of OmpR is inverted via cI lam so that light activity is being measured, instead of darkness. The luxI gene is expressed in the presence of light. Its gene product produces 3OC6HSL. Note that cI lam and luxI both have LVA degradation tags to ensure they don’t accumulate.<br> (c) The luxr gene is constitutively expressed. The gene product detects 3OC6HSL. When the HSL concentration is high enough, the lux pR promoter is activated, resulting in the production of ECFP. Note that ECFP has a degradation tag.]] |
| - | + | '''''B. Weight Cells''''' | |
| - | + | The weight cells control the amount of light permitted to pass by lacZ expression and cell death. When lacZ is expressed, less light passes; and when cell death occurs, any lacZ in the dead cell diffuses, allowing more light to pass. The delta rule (1) dictates how the weights should change. When there is input and a target output signal, the weight should increase. When there is input and output, the weight should decrease. Low heat will be used as the target output signal. Hence, when there is light and low heat, cell death will occur via suppression of the KanR gene, given the media contains kanamycin. When there is light and HSL molecules, lacZ will be produced. | |
| - | + | A light filter may be placed between the weight cells and the output cells to ensure the weight cells receive the maximum light in the on-state, while the output cells receive a degree of light that is closer to the on/off transition point. This way, the weight cells will have more effect on the output. | |
| + | The system will be designed so that the output cells and the weight cells do not share the same media. This way, the media from the output cells can be transferred to the weight cells to initiate network training once the HSL molecules have been generated and evenly distributed. | ||
| - | + | [[Image:fig4.png|frame|center|Fig. 4. Genetic parts for the weight cells.<br> (a) Light receptors are constitutively expressed. The activation of OmpR is inverted via tetR.<br> (b) In the presence of light, luxR and lac ts are expressed. The luxR gene product detects 3OC6HSL and activates lux pR. When lux pR is activated, lacZ is expressed. The lac ts gene product inhibits ampR expression when the temperature is reduced, resulting in cell death.]] | |
| - | + | '''''D. Training''''' | |
| - | + | This network can be trained to either allow one input to propagate to the output, or to function as a AND or OR gate. Training is performed by presenting the network with all input and target output combinations for the logic function desired of the network. A typical training run would proceed as follows: | |
| + | # ''Presentation of input to network:'' Light input is presented to the output cells for the predetermined exposure time. The weight cells will not have kanamycin or X-gal in the media to prevent blue color production or cell death. The output cells should have kanamycin and X-gal in the media since the media will be transferred to the weight cells for training. | ||
| + | # ''Blocking of output cells to further input:'' The light input should be blocked from reaching the output cells (but not the weight cells) in order to allow the system to reach steady state. | ||
| + | # ''Presentation of target output to weight cells:'' The appropriate temperature should now be set for the desired target output. | ||
| + | # ''Enabling of training mode:'' Training mode is enabled by transferring the media from the output cells to the weight cells. | ||
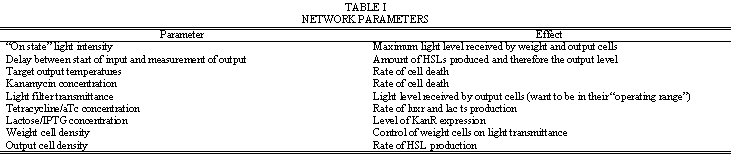
| - | + | Table I. is a list of parameters that can be adjusted to control how the network behaves. Note that for the same actual and target output levels, the weight cells should be “zeroed” so that the rates of lacZ production and cell death are such that their effects cancel each other with respect to light transmittance. | |
| - | + | [[Image:table1.jpg]] | |
| - | + | ||
| - | + | ||
| - | + | '''''E. Simulation''''' | |
| - | + | Simulation will need to model the internal chemistry of the cell, as well as the propagation of HSL signals outside the cell. This will aid in determining appropriate values for the network parameters listed in Table 1. | |
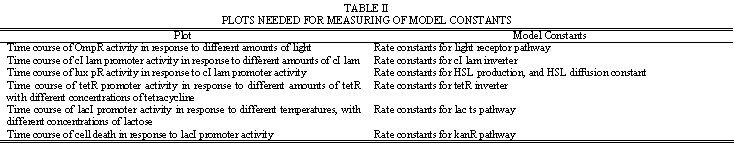
| + | The modeling process will first involve constructing a model of the basic neural network functionality. Once this is working as tested by hypothetical inputs and weight cell transmittances, full weight cell dynamics can be implemented. Table 2 provides a guideline on some of the experimental plots that will be needed to develop the model. | ||
| - | + | [[Image:table2.jpg]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
=== SIGNIFICANCE OF WORK === | === SIGNIFICANCE OF WORK === | ||
| - | Developing a NN using a genetic regulatory network would provide a stepping stone for the creation of biological systems that operate on fuzzy logic. These types of systems would have the ability to adapt to changing environmental conditions without the need to design traditional predicate logic based genetic circuitry. | + | Developing a NN using a genetic regulatory network would provide a stepping stone for the creation of biological systems that operate on fuzzy logic. These types of systems would have the ability to adapt to changing environmental conditions without the need to design traditional predicate logic based genetic circuitry. Moreover, by implementing a neural network using biological components, one can take advantage of the parallel processing capabilities of biological systems. |
| - | While relatively simple in functionality, this design will provide a basis for more complicated NN topologies | + | While relatively simple in functionality, this design will provide a basis for more complicated NN topologies. |
=== REFERENCES === | === REFERENCES === | ||
Latest revision as of 23:12, 26 October 2007
Contents |
Project: E. Coli Neural Network
INTRODUCTION
Neural networks (NNs) are nonlinear systems capable of distributed processing over a number of simple interconnected units. By adjusting connections between these units, NNs are capable of learning. Similar to NNs, a culture of Escherichia coli cells can be viewed as a distributed processing system with connections formed by intercellular signaling pathways. By engineering the cells to interact with each other in predetermined ways, it should be feasible to develop a bacteria based neural network.
OBJECTIVES
The goal of the proposed project is to develop a simple two-input-one-output feedforward NN using E. coli cells. This network will be capable of being trained to function as different types of digital logic gates.
REVIEW OF PERTINENT LITERATURE
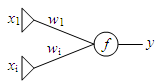
The neural network topology being examined in this proposal is known as a perceptron. Perceptrons are linear classifiers, which output the function of a linear combination of inputs [1]. The factor by which each input is multiplied is known as the weight. A perceptron is visually depicted as a layer of input units which are connected by network weights to a single output unit which performs a function on the weighted sum of the inputs (Fig. 1). For the proposed project, only one input (x1) will be used.
Training of feedforward neural networks typically employs a method known as backpropagation [2]. Backpropagation involves a forward pass where the output of the neural network is computed for a given set of inputs. A backward pass is made where network weights are adjusted based on the error between the actual output and the expected output. The delta rule is commonly employed to determine the amount of weight change. For perceptrons, a simplification of the delta rule can be made where the weight change is equal to (delta)wi = a xi (d-y) (1).
where d is the target output and alpha is a constant that controls the amplitude of the weight adjustment [1]. Training is done by exposing the network to a training set until the error between the actual and expected output decreases within a defined tolerance level.
Fig. 1. Generalized topology of a perceptron.
METHODS
A. Basic E. Coli Neural Network Structure
The input to the proposed network will be light and the output will be ECFP expression. Two red light sources will comprise the input nodes. The cell population will consist of output cells and weight cells. The output cells, which represent the output node, will be physically separated into three neighboring groups by a membrane to prevent cell migration. The first two groups detect light from the two network inputs. The weight cells will be mounted above two groups of output cells and will act as a light filter to control the amount of light received by the output cells. The weight cell groups will be separated via a membrane to prevent cell migration. The amount of light picked up by each individual output cell group is integrated via production of HSL signaling molecules. These molecules are picked up by all output cell groups. The third group of output cells has no weight cells above it and exists only for the measurement of ECFP expression. For each input-output measurement and between training sets, the media should be replaced to remove HSL and blue substrate molecules. An abstracted diagram of the topology is illustrated in Fig. 2. The genetic parts for the output cells are shown in Fig. 3.
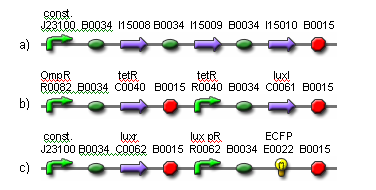
(a) The light receptor is constitutively expressed.
(b) OmpR is activated in darkness when the EnvZ domain on the light receptor is phosphorylated. The activation of OmpR is inverted via cI lam so that light activity is being measured, instead of darkness. The luxI gene is expressed in the presence of light. Its gene product produces 3OC6HSL. Note that cI lam and luxI both have LVA degradation tags to ensure they don’t accumulate.
(c) The luxr gene is constitutively expressed. The gene product detects 3OC6HSL. When the HSL concentration is high enough, the lux pR promoter is activated, resulting in the production of ECFP. Note that ECFP has a degradation tag.
B. Weight Cells
The weight cells control the amount of light permitted to pass by lacZ expression and cell death. When lacZ is expressed, less light passes; and when cell death occurs, any lacZ in the dead cell diffuses, allowing more light to pass. The delta rule (1) dictates how the weights should change. When there is input and a target output signal, the weight should increase. When there is input and output, the weight should decrease. Low heat will be used as the target output signal. Hence, when there is light and low heat, cell death will occur via suppression of the KanR gene, given the media contains kanamycin. When there is light and HSL molecules, lacZ will be produced.
A light filter may be placed between the weight cells and the output cells to ensure the weight cells receive the maximum light in the on-state, while the output cells receive a degree of light that is closer to the on/off transition point. This way, the weight cells will have more effect on the output. The system will be designed so that the output cells and the weight cells do not share the same media. This way, the media from the output cells can be transferred to the weight cells to initiate network training once the HSL molecules have been generated and evenly distributed.
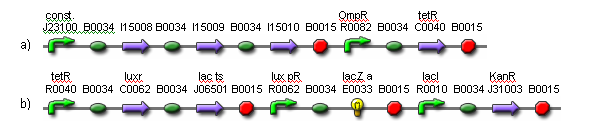
(a) Light receptors are constitutively expressed. The activation of OmpR is inverted via tetR.
(b) In the presence of light, luxR and lac ts are expressed. The luxR gene product detects 3OC6HSL and activates lux pR. When lux pR is activated, lacZ is expressed. The lac ts gene product inhibits ampR expression when the temperature is reduced, resulting in cell death.
D. Training
This network can be trained to either allow one input to propagate to the output, or to function as a AND or OR gate. Training is performed by presenting the network with all input and target output combinations for the logic function desired of the network. A typical training run would proceed as follows:
- Presentation of input to network: Light input is presented to the output cells for the predetermined exposure time. The weight cells will not have kanamycin or X-gal in the media to prevent blue color production or cell death. The output cells should have kanamycin and X-gal in the media since the media will be transferred to the weight cells for training.
- Blocking of output cells to further input: The light input should be blocked from reaching the output cells (but not the weight cells) in order to allow the system to reach steady state.
- Presentation of target output to weight cells: The appropriate temperature should now be set for the desired target output.
- Enabling of training mode: Training mode is enabled by transferring the media from the output cells to the weight cells.
Table I. is a list of parameters that can be adjusted to control how the network behaves. Note that for the same actual and target output levels, the weight cells should be “zeroed” so that the rates of lacZ production and cell death are such that their effects cancel each other with respect to light transmittance.
E. Simulation
Simulation will need to model the internal chemistry of the cell, as well as the propagation of HSL signals outside the cell. This will aid in determining appropriate values for the network parameters listed in Table 1. The modeling process will first involve constructing a model of the basic neural network functionality. Once this is working as tested by hypothetical inputs and weight cell transmittances, full weight cell dynamics can be implemented. Table 2 provides a guideline on some of the experimental plots that will be needed to develop the model.
SIGNIFICANCE OF WORK
Developing a NN using a genetic regulatory network would provide a stepping stone for the creation of biological systems that operate on fuzzy logic. These types of systems would have the ability to adapt to changing environmental conditions without the need to design traditional predicate logic based genetic circuitry. Moreover, by implementing a neural network using biological components, one can take advantage of the parallel processing capabilities of biological systems.
While relatively simple in functionality, this design will provide a basis for more complicated NN topologies.
REFERENCES
[1] O. Weisman, The Perceptron. [Online]. Available: http://www.cs.bgu.ac.il/~omri/Perceptron
[2] B. Bardakjian, Cellular Bioelectricity Course Notes. 2005.
Back to the Toronto team main page