UCSF/Main Introduction
From 2007.igem.org
< UCSF(Difference between revisions)
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | [[Image:GIntro1.png]] | + | {| |
| - | [[Image:GIntro2.png]] | + | |[[Image:GIntro1.png]]|| '''This year’s UCSF iGEM team was truly a collaborative effort between several institutions. Because UCSF has no undergraduate program, we thought it was an excellent opportunity to extend the educational power of iGEM by partnering up with a local high school. Five of our iGEM team members come directly from San Francisco’s Abraham Lincoln High School and are graduates of a intensive biotechnology program taught by George Cachianes and Julie Reis. Two other students, one from Palo Alto High School and one from UC Berkeley, also make up the team. Because of the unique team composition, our program focused on communicating core concepts in research science and synthetic biology while also actively pursuing the iGEM challenge. As a result, our summer was spent exploring synthetic biology through creative, educational, experimental and interactive angles.''' |
| - | [[Image:GIntro3.png]] | + | |- |
| - | [[Image:GIntro4.png]] | + | |[[Image:GIntro2.png]]|| '''Through our summer’s work, a conceptual focus resolved: we wanted to address the significance of space within cells. As synthetic biologists, we frequently on engineering biological systems by simply introducing novel functionalities. Rarely do we concern ourselves with myriad roles space can play on cellular behavior. As such, our team sought to address the significant of spatial localization by directing biology through synthetic assemblies and organelles. ''' |
| - | [[Image:Gintro5.png]] | + | |- |
| - | [[Image:Gintro56.png]] | + | |[[Image:GIntro3.png]]||'''The following statement will probably raise few, if any objections: the processes that cells carry out are complex and many. To harness energy, maintain homeostasis, or respond to environmental cues, cells must execute numerous ordered, multi-step and often interrelated pathways. As such, doesn't seems like a logistical nightmare to have the enzymes, substrates and intermediates for all of these pathways freely diffusing around in the cytoplasm? This task becomes even more daunting as one thinks beyond bacteria and begins thinking about the complexity of eukaryotes.''' |
| - | [[Image:Gintro7.png]] | + | |- |
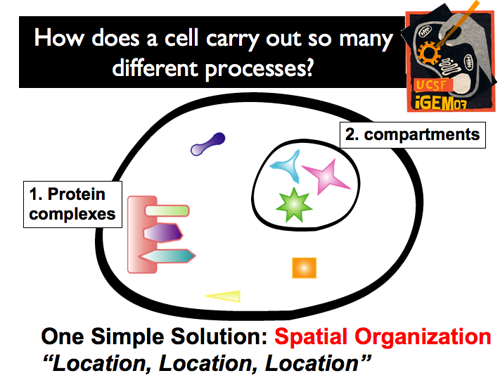
| - | [[Image:Gintro8.png]] | + | |[[Image:GIntro4.png]]|| '''Of course, cells have evolved to avoid this potential traffic jam. What’s more, they’ve done so with a very simple solution: spatial organization. By selectively localizing and organizing particular cellular components in different regions, messages can be quickly and accurately sent, molecules can be correctly synthesized, etc. This spatial localization can be achieved through two different mechanisms: the formation of protein complexes (which organize and regulate) and compartments (which physically isolate an interior environment from the cytoplasm).''' |
| + | |- | ||
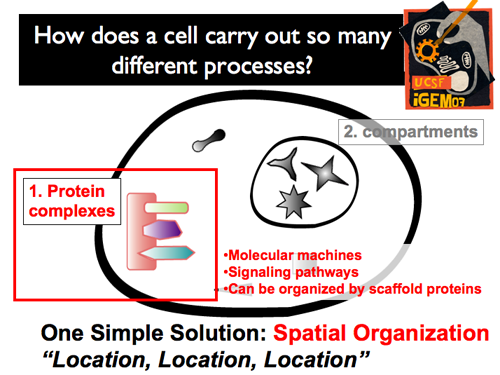
| + | |[[Image:Gintro5.png]]||'''Protein complexes can specifically control the wiring of a cellular circuit by regulating both its composition and its connectivity. This simple approach can create powerful results. Frequently, proteins are organized upon scaffold proteins, where multiple, specific docking sites specifically recruit interacting partners. This prevents cross talk while also executing multi-step tasks efficiently.''' | ||
| + | |- | ||
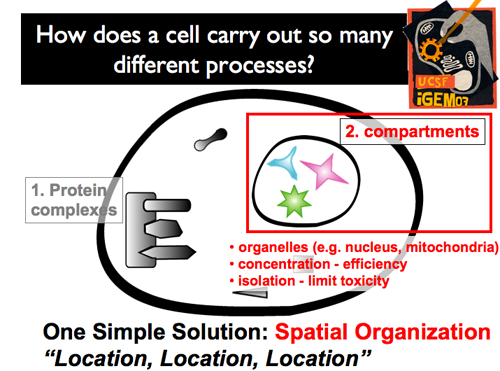
| + | |[[Image:Gintro56.png]]|| '''Distinct, membrane-bound compartments within a cell also offer one ultrapowerful advantage: the 'business' of the interior does not interfere with 'business' of the cytoplasm (and vice-versa). Unique environments can be created within (pH, unique concentrations). Higher local concentrations can be easily achieved by the benefit of a reduced volume. Moreover, Controlling what’s inside and what’s outside can create specific protein-protein interactions. As a result, cells can power specific circuits, functioning in specific environments. And, the compartmental membrane can either insulate this circuit from the outside environment, or insulate the outside environment from this new circuit.''' | ||
| + | |- | ||
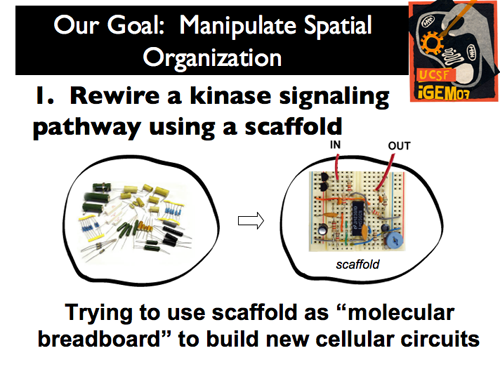
| + | |[[Image:Gintro7.png]]|| '''The power of space within the cell has not yet been harnessed by synthetic biology. As such, our iGEM project approached the manipulation of spatial organization through two distinct projects. First, by treating the scaffold as a “molecular breadboard,” we sought to rewire a kinase signaling pathway to build new cellular circuits.''' | ||
| + | |- | ||
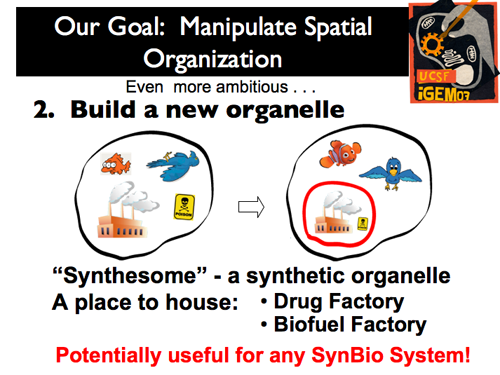
| + | |[[Image:Gintro8.png]]|| '''Our second and more ambitious project was to create a stable membrane compartment within a eukaryote: we wanted to create a new organelle (or a “synthesome,” as we call it). After this is accomplished, the options are nearly limitless for its function! Two very relevant applications are the use of the new organelle as a Drug or Biofuel factory.''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div style="text-align: right;"> <font size=4><font color=firebrick>Click '''[https://2007.igem.org/UCSF/Intro here]''' to learn more about our first project!'''</font></font></div> | ||
Latest revision as of 23:48, 28 September 2007
Click here to learn more about our first project!