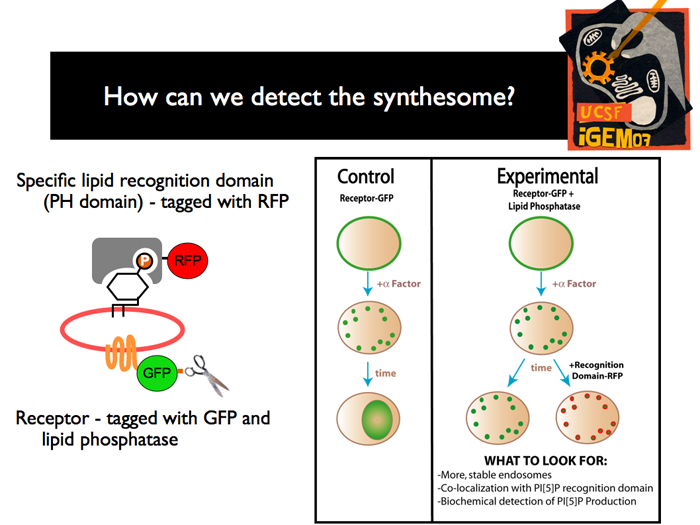
 | Once we have created all of the constructs and yeast strains required, we have to confirm the creation of the synthetic organelle. We plan to use two main methods: The first is to use flourescence microscopy. The receptor that is endocytosed is fused to GFP, and a “Plekstrin Homology” (PH) domains specific for PI[5]P is tagged with RFP. In the control strain, before induction, we should see a ring of green flourescence, representing the membrane associated receptors. After induction, there should be specks of green, indicating that the receptor has been endocytosed. Finally, we should see a green glow from the vacuole, as the endosomes are degraded. In the experimental strain, before induction, we should also see membrane associated receptor. After induction, we will also see endocytosis, but after some time has passed, we should be able to see GFP/RFP localization, in specks around the cell, indicating the existence of 5’ phosphoinositides lining the membrane of new organelles. Radioactivity techniques will be also used to detect the production of PI[5]P.
|