USTC/SuXiaofeng
From 2007.igem.org
Xiaofeng Su (Talk | contribs) |
Xiaofeng Su (Talk | contribs) (→Undergruduate Student of Cellular and Molecular Biology,USTC) |
||
| (49 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | [[Image:USTC_ALLEN.jpg|thumb|left| | + | [[Image:USTC_ALLEN.jpg|thumb|left|Xiaofeng Su(Allen Su)-Undergraduate of Cellular and Molecular Biology in School of Life Sciences, USTC, P.R. China]] |
==XIAOFENG SU== | ==XIAOFENG SU== | ||
=====Undergruduate Student of Cellular and Molecular Biology,USTC===== | =====Undergruduate Student of Cellular and Molecular Biology,USTC===== | ||
| - | Email:allensue@mail.ustc.edu.cn (preference) | + | '''Email:''' [mailto:allensue@mail.ustc.edu.cn allensue@mail.ustc.edu.cn] (preference) or [mailto:xiaofsu@gmail.com xiaofsu@gmail.com] |
| + | |||
| + | '''Phone:''' +86-551-3602469 (Lab) | ||
| + | |||
| + | '''Mobile:''' +86-13877561295 | ||
| + | |||
| + | '''Address:''' Room 439, School of Life Sciences, USTC, Hefei, Anhui, P.R.China, 230026 | ||
| + | |||
| + | =====Research Interest===== | ||
| + | |||
| + | *Directed Evolution for Seeking New Protein-DNA Interactions | ||
| + | |||
| + | *Approaches of Synthetic Biology for Forming Novel "Genetic Engineering Machines" | ||
| + | |||
| + | *Differentiation and Development of Stem Cells | ||
| + | |||
| + | '''Research Work''' | ||
| + | |||
| + | *;Overall Description | ||
| + | |||
| + | Unlike the real wires of electrocircuit board, in cytoplasm, chemical signaling molecules in an relatively open systems. For obtaining the signaling transduction parts of repression with high fidelity, I've experimentally designed and acquired some specific repressor-promoter pairs(R-P pairs or P-R pairs)based on Lactose Operon by directed evolution on plate. Besides, through quantitative assay,the novel artificial R-P pairs I selected have been tested for their binding performance so that P-R pairs of highest affinity and specificity can make a figure out of P-R pair candidates. Most of the parts in my work have been BioBricks-Standardized and work as BioBrick parts. | ||
| + | |||
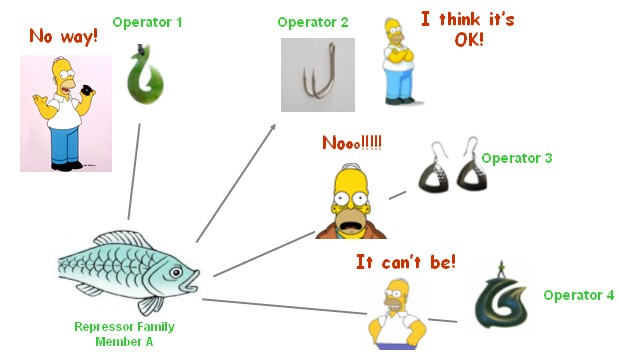
| + | [[Image:USTC_allen1.jpg|thumb|center|500px|'''Figure.1.'''My project description by visual comics: All we need are the specific artificial 'repressor fish' that can definitely bite the specific 'operator hook' exclusively]] | ||
| + | |||
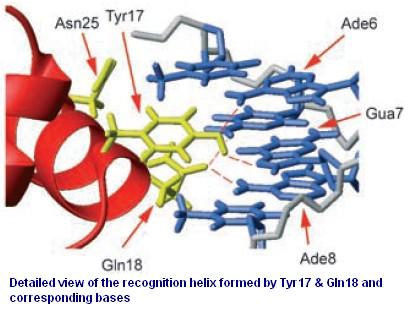
| + | [[Image:USTC_allen2.jpg|thumb|right|200px|'''Figure.2.''' My Work Basis-By Redesigning lac-repressor and operator. This figure comes from 'Roberto Kopke Salinas, etc.' ChemBioChem 2005, 6, 1628 – 1637]] | ||
| + | |||
| + | *;Experimental Design | ||
| + | #Construction of Expression Library of Lac-Repressor Family [Collaboration] | ||
| + | #Synthesis of Promoter Sequence with Specific Operators | ||
| + | #Construction of Low-copy Reporter System with Specific Opertors | ||
| + | #Selection of Promoter-Repressor Pair(P-R Pair) including re-testing validity of these combination | ||
| + | #Transfering the Operators to Double Reporter Systems [Collaboration] | ||
| + | #Quantitative assay of Repression Intensity and Specificity of P-R Pairs | ||
| + | #Results From RM(Repression Matrix) to ORM(Orthogonal RM) | ||
| + | |||
| + | |||
| + | *;Work Process and Summary | ||
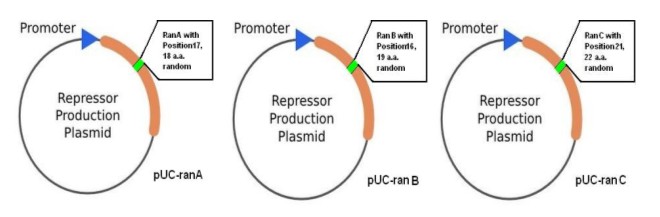
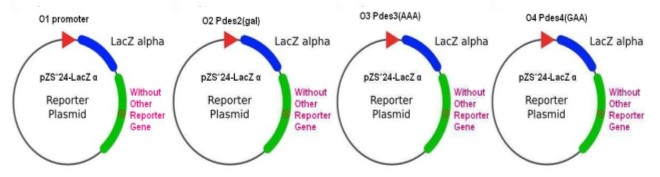
| + | '''(A)'''The Directed Evolution must be conducted on the base of 2 components-the repressor system and promoter-reporter system.By collaborations with other team members, I've successfully establish these two systems shown as below.[[Image:USTC_allen3.jpg|thumb|left|300px|'''Figure.3.'''Three kinds of plasmids birthing random mutated repressor called repressor system]][[Image:USTC_allen4.jpg|thumb|center|380px|'''Figure.4.'''Four kinds of plasmids with different specific promoters containing lacZalpha as reporter gene]] | ||
| + | |||
| + | |||
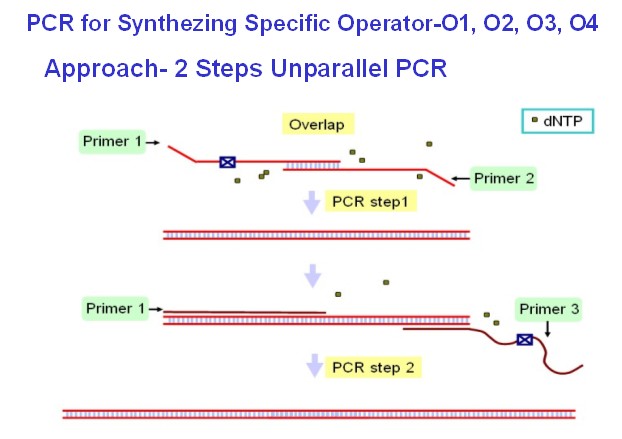
| + | '''(B)'''Within the work of establishment of two selection systems, a time consuming step is synthesis of target specific promoters that contain the mutated site design by Do. On account of economic and convenient aspect, I use 2-Step unparallel PCR by 3 fragments of primers and 2 times of PCR with different reaction conditions.As Below. | ||
| + | |||
| + | [[Image:USTC_allen5.jpg|thumb|left|320px|'''Figure.5.'''The PCR plan for synthesis of specific promoters]][[Image:USTC_allen6.jpg|thumb|center|300px|'''Figure.6.'''The PCR Results]] | ||
| + | |||
| + | |||
| + | |||
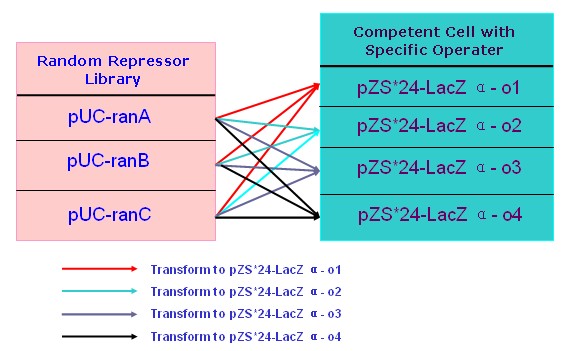
| + | '''(C)'''I've develop the selection work by the means of Blue White Selection on top agar Luria-Bertani broth to obtain the R-P pairs.Below are some pictures from my experiment design and experiment work. | ||
| + | |||
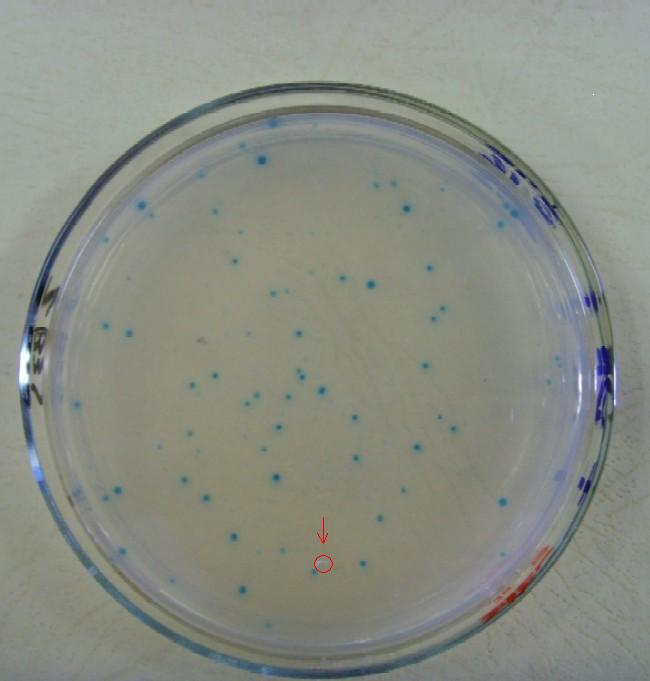
| + | [[Image:USTC_allen10.jpg|thumb|left|300px|'''Figure.7.'''My arranged selection work desciption]] [[Image:USTC_allen7.jpg|thumb|right|200px|'''Figure.8.'''Selection Results-the Red-Mark the colony are a target]] | ||
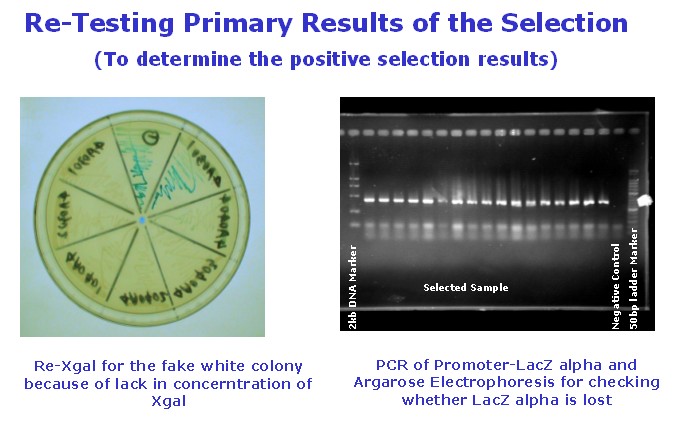
| + | [[Image:USTC_allen8.jpg|thumb|center|300px|'''Figure.9.'''retest of my results of P-R pairs]] | ||
| + | |||
| + | |||
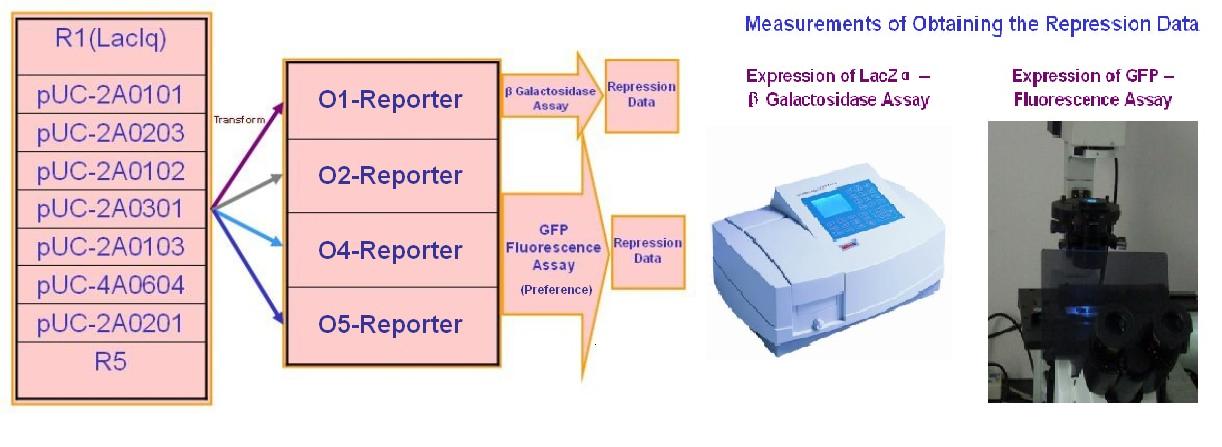
| + | '''(D)'''I've conducted a 'cross repression' test by transforming the selected repressors to their specific promoter-reporter system and other non-specific promoter-reporter systems.We selected 7 repressor-promoter pair candidates from Blue/White Screening results above for quantitive assay of specificity and affinity. In addition, 2 existed represor-promoter pairs are added to this work as new candidates. Then, each repressor-expression plasmid is transform to each Top10 competent cell with specific target promoters. Eventually, each expression quantity of LacZ alpha or GFP is measured by ONPG assay(LacZ) or fluorescent assay(GFP).The process are shown as below. | ||
| + | |||
| + | [[Image:USTC_ crossrepressiontest.jpg|center|600px]] | ||
| + | |||
| + | *;Key Results | ||
| + | |||
| + | The consequent data of reporter's expression is, by formula, converted to the Repression Value(R.V.) representing the repression intensity of each repressor-promoter pair. From the formula below we can see higher value of R.V. represents the higher expression of reporter gene and indicates a lower repression while the reverse represents a high repression. The sets of R.V. is depicted on the scheme below which have been transformed to corresponding Repression Matrix-more visual that the coordinate scheme. And the Orthogonal Repression Matrix birthing from Repression Matrix can be used to acquire specific repressor-promoter pairs with high specificity. | ||
| + | |||
| + | [[Image:USTC_rv.jpg|500px|center]] | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | === Repression Scheme === | ||
| + | |||
| + | The directed results are charted on one coordinate scheme. | ||
| + | |||
| + | [[Image:USTC_RepressionScheme.jpg|thumb|center|550px|'''Figure 7''' This scheme indicates the cross repression test of our repressor and promoter candidates. The vertical axis tell us the Repression Value(R.V.) of each repressor-promoter pairs by plots, in which the higher R.V.shows the higher expression of reporter protein(even exceed the condition without repressors)-the low repression. The horizontal axis represents the different repressors, and the colors represent different promoters]] | ||
| + | --> | ||
| + | |||
| + | '''a) Repression Matrix ''' | ||
| + | |||
| + | Performances of our wires are shown as so-called "Repression Matrix", an array of R.V. with variant combinations of artificial repressors and operators. A "Repression Matrix" taken from the literature and uniformed is also plotted in [[USTC/RepressionMatrixFromLiterature|this page]]. | ||
| + | |||
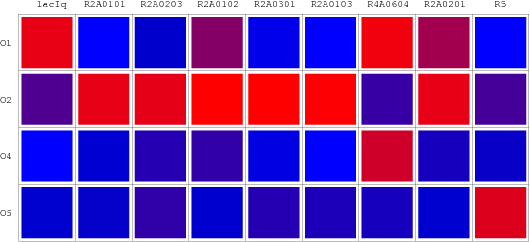
| + | [[Image:USTC_RepressionMatrix.png|thumb|center|500px|'''Figure 10''' Repression Metrix(RM):The repression matrix reveals the binding affinity of different repressor candidates with various specific promoters by different colors. The deeper the red will be, the higher the repression will appear. While the lighter the blue is the weaker the repression will show.]] | ||
| + | |||
| + | '''b)Orthologal Repression Matrix''' | ||
| + | |||
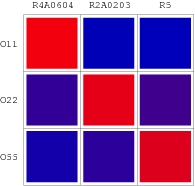
| + | [[Image:USTC_OrthologalRepressionMatrix.png|thumb|rught|200px|'''Figure 11''' Orthologal Repression Metrix(ORM)-One of the ORM comes from above RM:The red diagonal of this matrix indicates that each target repressor can only firmly bind to its specific target promoter and has weak or even no binding to other operator sequences.]] | ||
| + | |||
| + | Several 'wires' without interference are selected based on that repressors can only specifically bind to the right promoter without strong repressed interaction with other promoters which have their own special repressors. Thereby, the red color plot in Repression Matrix should be placed on the right site by interchange of columns so that each row and each column in the Matrix can have only one red plot. The other repressor candidate columns which cannot pass muster will be deleted from the RM. Eventually, a 3x3 Orthogonal Repression Matrix for [[USTC/Demonstration|the demonstration system]] shown in Figure 9, which looks like an orthogonal matrix in mathematics, contains single red plot in each column & each row, reflecting the specific repression. | ||
| + | |||
| + | *;Repressor Design in silico | ||
| + | |||
| + | Besides my artificial design work in experiment, we also conduct a program for screening the target repressor-promoter pairs via computational biology mainly by Ding Bo, shown as the following website: [[Repressor Design in silico]] | ||
| + | |||
| + | *;[[Reference]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | *;Difficulties & Overcome | ||
Latest revision as of 13:10, 24 August 2009
XIAOFENG SU
Undergruduate Student of Cellular and Molecular Biology,USTC
Email: allensue@mail.ustc.edu.cn (preference) or xiaofsu@gmail.com
Phone: +86-551-3602469 (Lab)
Mobile: +86-13877561295
Address: Room 439, School of Life Sciences, USTC, Hefei, Anhui, P.R.China, 230026
Research Interest
- Directed Evolution for Seeking New Protein-DNA Interactions
- Approaches of Synthetic Biology for Forming Novel "Genetic Engineering Machines"
- Differentiation and Development of Stem Cells
Research Work
- Overall Description
Unlike the real wires of electrocircuit board, in cytoplasm, chemical signaling molecules in an relatively open systems. For obtaining the signaling transduction parts of repression with high fidelity, I've experimentally designed and acquired some specific repressor-promoter pairs(R-P pairs or P-R pairs)based on Lactose Operon by directed evolution on plate. Besides, through quantitative assay,the novel artificial R-P pairs I selected have been tested for their binding performance so that P-R pairs of highest affinity and specificity can make a figure out of P-R pair candidates. Most of the parts in my work have been BioBricks-Standardized and work as BioBrick parts.
- Experimental Design
- Construction of Expression Library of Lac-Repressor Family [Collaboration]
- Synthesis of Promoter Sequence with Specific Operators
- Construction of Low-copy Reporter System with Specific Opertors
- Selection of Promoter-Repressor Pair(P-R Pair) including re-testing validity of these combination
- Transfering the Operators to Double Reporter Systems [Collaboration]
- Quantitative assay of Repression Intensity and Specificity of P-R Pairs
- Results From RM(Repression Matrix) to ORM(Orthogonal RM)
- Work Process and Summary
(B)Within the work of establishment of two selection systems, a time consuming step is synthesis of target specific promoters that contain the mutated site design by Do. On account of economic and convenient aspect, I use 2-Step unparallel PCR by 3 fragments of primers and 2 times of PCR with different reaction conditions.As Below.
(C)I've develop the selection work by the means of Blue White Selection on top agar Luria-Bertani broth to obtain the R-P pairs.Below are some pictures from my experiment design and experiment work.
(D)I've conducted a 'cross repression' test by transforming the selected repressors to their specific promoter-reporter system and other non-specific promoter-reporter systems.We selected 7 repressor-promoter pair candidates from Blue/White Screening results above for quantitive assay of specificity and affinity. In addition, 2 existed represor-promoter pairs are added to this work as new candidates. Then, each repressor-expression plasmid is transform to each Top10 competent cell with specific target promoters. Eventually, each expression quantity of LacZ alpha or GFP is measured by ONPG assay(LacZ) or fluorescent assay(GFP).The process are shown as below.
- Key Results
The consequent data of reporter's expression is, by formula, converted to the Repression Value(R.V.) representing the repression intensity of each repressor-promoter pair. From the formula below we can see higher value of R.V. represents the higher expression of reporter gene and indicates a lower repression while the reverse represents a high repression. The sets of R.V. is depicted on the scheme below which have been transformed to corresponding Repression Matrix-more visual that the coordinate scheme. And the Orthogonal Repression Matrix birthing from Repression Matrix can be used to acquire specific repressor-promoter pairs with high specificity.
a) Repression Matrix
Performances of our wires are shown as so-called "Repression Matrix", an array of R.V. with variant combinations of artificial repressors and operators. A "Repression Matrix" taken from the literature and uniformed is also plotted in this page.
b)Orthologal Repression Matrix
Several 'wires' without interference are selected based on that repressors can only specifically bind to the right promoter without strong repressed interaction with other promoters which have their own special repressors. Thereby, the red color plot in Repression Matrix should be placed on the right site by interchange of columns so that each row and each column in the Matrix can have only one red plot. The other repressor candidate columns which cannot pass muster will be deleted from the RM. Eventually, a 3x3 Orthogonal Repression Matrix for the demonstration system shown in Figure 9, which looks like an orthogonal matrix in mathematics, contains single red plot in each column & each row, reflecting the specific repression.
- Repressor Design in silico
Besides my artificial design work in experiment, we also conduct a program for screening the target repressor-promoter pairs via computational biology mainly by Ding Bo, shown as the following website: Repressor Design in silico
- Difficulties & Overcome