Virginia/Projects/1
From 2007.igem.org
(→'''Biosynthesis of butanol biofuel by a synthetic metabolic pathway in ''E. coli''''') |
(→Biobrick and pathway design) |
||
| Line 59: | Line 59: | ||
[[Image:untitled.jpg]] | [[Image:untitled.jpg]] | ||
| - | This is the pathway we are incorporating into E.Coli. | + | This is the pathway we are incorporating into E.Coli. |
| + | |||
| + | Clostridium acetbutylicum has been known to produce butanol anaerobically in nature. For our project we will be | ||
| + | utilizing two butanol sythesis pathways available to us from its genome. In addition to this, we plan to provide a | ||
| + | carbon source for butanol biosythesis by importing a cellulase gene from Saccaruphagus degradans, and we plan to | ||
| + | facilitate the proton gradient required for ATP sythesis through the use of a proteorhodopsin system. | ||
| + | |||
| + | The diagram below depicts the complete system that we plan to create. The added pathways are: 3, 4, 5, 6, 9, 10, 11. Excluded from this image is our plans for the cellulose digestion pathway, which essentially feed into the production of pyruvate. | ||
The specific proteins we need the cells to produce for this pathway are: | The specific proteins we need the cells to produce for this pathway are: | ||
Revision as of 18:29, 19 July 2007
Biosynthesis of butanol biofuel by a synthetic metabolic pathway in E. coli
We have designed a metabolic system capable of cellulose degradation and light metabolism in order to power the biosynthesis of butanol fuel. This hybrid molecular engine is built from standard biological parts and may be used in future designs in order to drive forward cellular chemistry.
The coming years are going to require us to revamp our notions of a fuel economy. Our team hopes to show how synthetic biology can aid in tackling real-world problems not only to aid in the development of new fuel technologies, but also to help support the synthetic biology community by showing it's utility.
Background, Motivation, and References
Why Butanol?
As energy demands increase, the need for alternative fuel sources increases dramatically. US market size for butanol: 370 million gallons per year at a price of about $3.75 per gallon. That’s $1.4 billion.
What is it used for?
Chemical and textile solvent, organic synthesis, chemical intermediate, paint thinner, base of perfumes, and, most importantly, as a biofuel.
As a Biofuel
Butanol biofuel can be used in cars without making engine modifications. It produces more power than ethanol and almost as much power as gasoline.
Butanol better tolerates water contamination and is less corrosive than ethanol and more suitable for distribution through existing pipelines for gasoline.
Why isn’t it more widely spread?
Historically low yields and low concentrations of biobutanol when compared to bioethanol have prevented industry from having stronger interest.
Product tolerance is the main issue. Butanol-producing bacteria (Clostridia acetobutylicum) become limited in growth at approximately 2.5% concentration. Isolating the product at this concentration is not economical.
In the 1950s butanol production shifted from fermentation to being petrochemically-derived. This method continues to be the most popular today.
There are developments in biobutanol production, however. Recently BP and Dupont announced the conversion of an ethanol fermentation facility in the UK to a dedicated biobutanol plant. Biobutanol from this plant will be introduced in 2007.
References:
http://www.greencarcongress.com/2006/06/bp_and_dupont_t.html
http://i-r-squared.blogspot.com/2006/05/bio-butanol.html
http://en.wikipedia.org/wiki/Butanol
Our Project
Our goal is to isolate the pathway of butanol production existing in various organisms and engineering the metabolic pathway of E.Coli to produce butanol. Butanol limits bacterial growth by degrades cellular membranes, so the first step is to convey butanol tolerance to E.Coli. This will be accomplished via the use of tolerance genes from other bacterial species.
Next, we will transform the cells with the necessary enzymes for butanol production. These are explained in detail below. By growing the cells in anaerobic conditions and analyzing their product, we hope to tweak the pathway to produce maximum amounts of butanol.
One approach to this is to vary the energy sources the bacteria can use. By inserting genes coding for cellulase, we hope to give our cells the ability to use cellulose as an energy source. Agricultural waste would then become the feed for our strains. Additionally, the use of proteorhodopsin to supplement ATP production is planned. Proteorhodopsin allow the cells to harness light energy independent of oxygen in the environment and drive cellular metabolism.
Our final goal is to design a system that will allow E.Coli to be tolerant to butanol, produce butanol, and do so by exploiting various energy sources to increase efficiency and large-scale feasibility.
Biobrick and pathway design
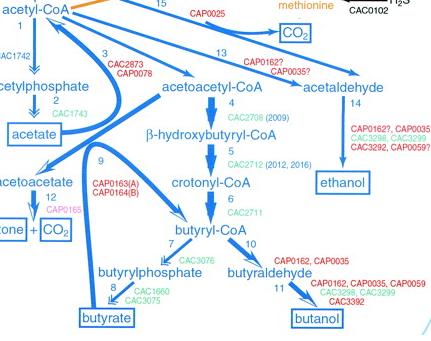
This is the pathway we are incorporating into E.Coli.
Clostridium acetbutylicum has been known to produce butanol anaerobically in nature. For our project we will be utilizing two butanol sythesis pathways available to us from its genome. In addition to this, we plan to provide a carbon source for butanol biosythesis by importing a cellulase gene from Saccaruphagus degradans, and we plan to facilitate the proton gradient required for ATP sythesis through the use of a proteorhodopsin system.
The diagram below depicts the complete system that we plan to create. The added pathways are: 3, 4, 5, 6, 9, 10, 11. Excluded from this image is our plans for the cellulose digestion pathway, which essentially feed into the production of pyruvate.
The specific proteins we need the cells to produce for this pathway are:
Specific cellulase,
butanol tolerance genes,
thiolase,
beta-hydroxybutyryl-CoA dehydrogenase,
crotonase,
butyryl coa dehydrogenase
AAD/AAD2
alcohol dehydrogenase,
AOTC/AOTD
We have designed the following biobricks:
Need this info.
Methods and Materials
We will analyze butanol production via gas chromatography and mass spectroscopy.
Our experimental scaffold (still in development) is as follows:
What is the butanol tolerance of WT cells and of transformed cells?
1) Butanol Tolerance
Hypothesis: Transformed cells will be more resistant to extracellular butanol than WT
* Add varying amounts of butanol to broths of transformed and non-transformed cells * Determine amounts of butanol cells can withstand before dying * Count cells with hemocytometer and blue stain
Note: Depending on butanol tolerance we will consider butanol tolerance transformed cells as wildtype (WT). If it doesn't give any significant tolerance we will just use the original strain.
Can cells survive with only cellulose carbon source?
2) Effect of Cellulase
Hypothesis: Transformed cells should be able to grow on low glucose/high cellulose media. No growth for WT.
* Transform cells with cellulase * Grow transformed cells and control cells on low glucose media supplemented with cellulose * Observe growth amounts
Does thiolase increase the production of acetoacetyl-CoA from acetyl-CoA relative to WT?
3) Effect of Thiolase
Hypothesis: Thiolase-transformed cells will have increased acetoacetyl-CoA production than WT, and most if cellulase-transformed
* Transform cells with thiolase * Transform cellulase-cells (ex.2) with thiolase * Plate transformed cells (thiolase and thiolase + cellualse, and control cells) * Test for acetoacetyl-CoA production
Note: If thiolase confers much greater acetoacetyl-CoA production, we will include it by default in the rest of the experiments requiring the central pathway.
How much butanol is produced from acetyl coa to butanol, with and without alcohol dehydrogenase?
4a) Acetyl CoA to Butanol (-cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol. WT will not
* Transform cells with thiolase, central pathway, aad/aad2 * Grow transformed and control cells * Test for Butanol
4b) Acetyl CoA to Butanol (-cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol, more than in ex. 4a. WT will not
* Transform cells with thiolase, central pathway, aad/aad2, alcohol dehydrogenase * Test for butanol production
How much butanol is produced from cellulase to acetyl coa to butanol, with and without alcohol dehydrogenase?
5a)Cellulase to Acetyl CoA to Butanol (+cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol. WT will not.
* Transform cells with cellulase, thiolase, central pathway, and aad/aad2 * Grow transformed and control cells * Test for Butanol
5b) Cellulase to Acetyl CoA to Butanol (+cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol, and at higher amounts than in ex. 5a. WT will not.
* Transform cells with cellulase, thiolase, central pathway, aad/aad2, alcohol dehydrogenase * Grow transformed and control cells * Test for Butanol
How much butanol is produced via only the bottom pathway (AOTC/AOTD genes) with and without the alcohol dehydrogenase?
6a)Butyrate (butyric acid) to Butanol (-cellulase, -thiolase, -central pathway, +aad, -alcohol dehydrogenase, +AOTC/AOTD) IN FLOW
Hypothesis: Pathway will produce butanol. There will be no butanol in WT
* Transform cells with AOTC/AOTD and aad/aad2 * Grow transformed and control cells * Measure butanol production with varying concentrations of initial butyric acid
6b) Butyrate (butyric acid) to Butanol (-cellulase, -thiolase, -central pathway, +aad, +alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: Pathway will produce butanol, more than in ex.6a. There will be no butanol in WT
* Transform cells with aad, AOTC/AOTD, and alc. dehydro. * Grow transformed and control cells * Measure butanol production with varying concentrations of initial butyric acid
How much butanol from acetyl coa to butanol, including AOTC/AOTD, and with and without alcohol dehydrogenase?
7a) Acetyl CoA to Butanol with Butyrate pathway (-cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway, but without alcohol dehyd. High butanol production expected.
* Transform cells with thiolase, central pathway, aad, AOTC/AOTD * Test for butanol production
7b) Acetyl CoA to Butanol with Butyrate pathway (-cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway, without cellulase. High butanol production expected.
* Transform cells with thiolase, central pathway, aad, AOTC/AOTD, and alc. dehydro. * Test for butanol production
How much butanol from cellulase to acetyl coa to butanol, including ctfa/b, and with and without alcohol dehydrogenase?
8a) Cellulase to Butanol with Butyrate pathway (+cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway, without alcohol dehydr. There should be maximum production of butanol.
* Transform cells with cellulase, thiolase, central pathway, aad, AOTC/AOTD, and alc. dehydro. * Test for butanol production
8b) Cellulase to Butanol with Butyrate pathway (+cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway. There should be maximum production of butanol.
* Transform cells with cellulase, thiolase, central pathway, aad, AOTC/AOTD, and alc. dehydro. * Test for butanol production
Will cells transformed with the proteorhodopsin pathway survive with only light, will it produce butanol, and if so how much will it increase butanol production when combined with other pathways?
9a) Effect of Proteorhodopsin
* Transform cells with proteorhodopsin genes * grow on minimal nutrient media * observe growth vs. control strains
9b) Proteorhodopsin to butanol
* If ex.9a cells grow at all, * transform cells with central pathway genes and proteorhodopsin * test for butanol production
9c) Proteorhodopsin to butanol, full pathway
* add proteorhodopsin to ex.8b cells * test for butanol production
Results and Conclusions
TBD