Virginia/Projects/3
From 2007.igem.org
| HOME | PROJECT INTRO | APPROACH | PROCEDURES | RESULTS | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Virginia BIOBRICKS] | [http://openwetware.org/wiki/IGEM:VGEM/2007/Notebook eNOTEBOOK] | [http://www.seas.virginia.edu/VGEM/ WEBSITE] |
Harvesting Cellulose and Light to Power Butanol Biosynthesis
Send comments to George McArthur
Results
Of the ten BioBricks that were designed, only proteorhodopsin (BBa_I711040) was propagated by E. coli. Any problems with digesting the construction vectors or inserts (i.e., the synthetic genes), ligating the two together, or transforming E. coli may have resulted in these failed attempts. However, it is likely that the gene products are somehow toxic to E. coli. Using a high copy number plasmid and a strong promoter, the high expression of these genes may have been lethal to the transformed cells. This is a strong possibility because DNA2.0 had trouble propagating many of the genes and ended up shipping PCR product instead.
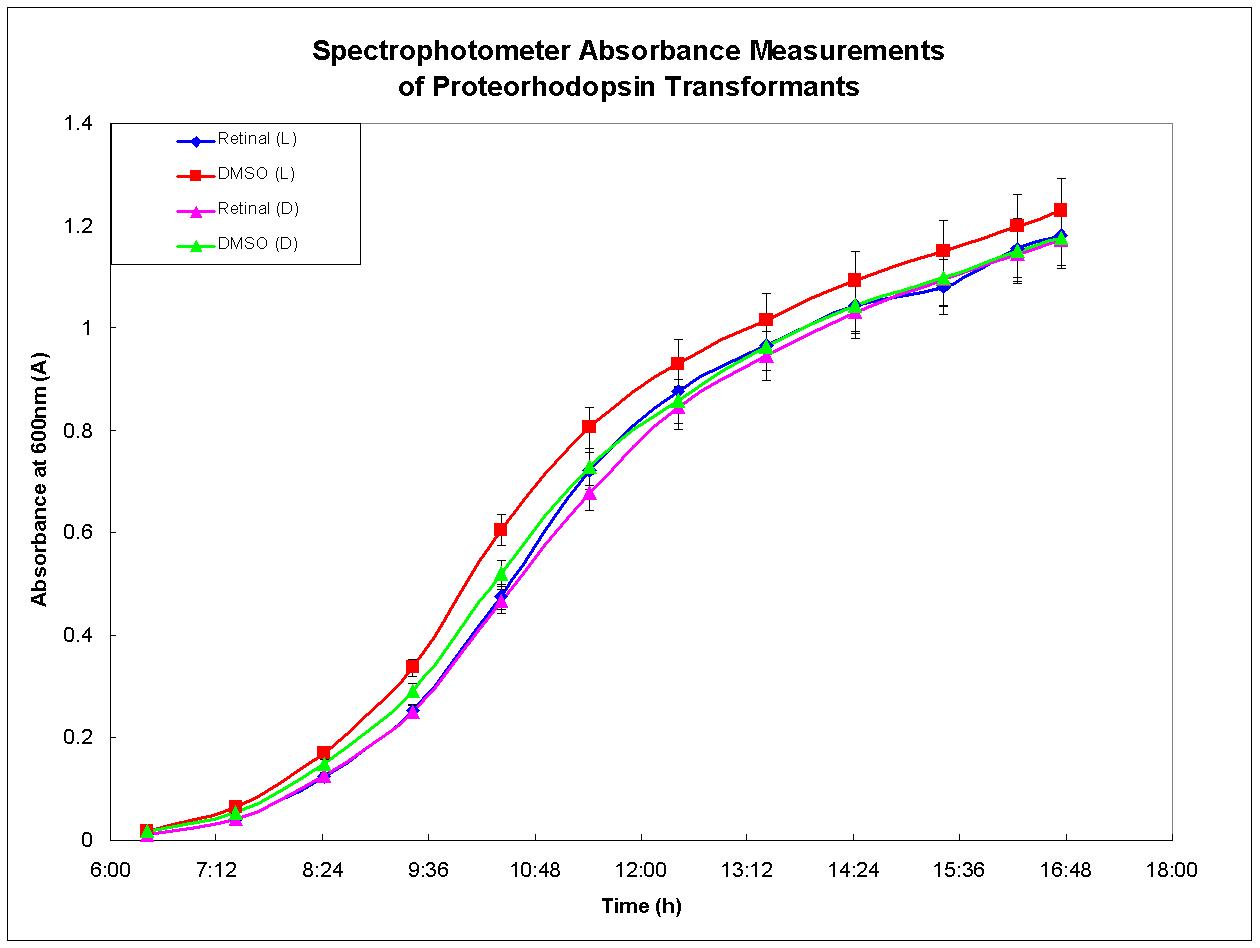
Proteorhodopsin-expressing bacteria were grown under optimal conditions. Four separate cultures were grown in LB broth. The first culture was grown in the presence of light and had retinal, required for functional proteorhodopsin, dissolved in DMSO added to the broth. There was no retinal added to the second culture, only DMSO. The third culture was grown under the same conditions as the first except that it was shielded from light. The fourth culture was also grown in the dark, but otherwise in the same conditions as the second culture. Figure 3, below, depicts the growth results for each culture. There was no growth advantage displayed by the culture grown in the light with retinal in the media; each culture behaved similarly. This experiment was performed again and similar results were observed. These results agree with past experiments developed by other researchers who have determined that proteorhodopsin is functional only when the cell is under respiratory stress (Walter et al., 2006). Therefore, proteorhodopsin may still be useful in the design of anaerobic metabolic pathways such as the butanol biosynthesis pathway.
Although the putative “butanol tolerance” gene was not propagated, butanol tolerance via directed evolution was attempted. Butanol tolerance tests were performed by growing cells up in LB broth with different concentrations of butanol and then transferring the surviving cells from the highest butanol concentration to fresh LB with different butanol levels. This technique proved inadequate during the short time frame which we were able to perform experiments. Ideally, we would have grown the cells up between 500 and 1000 generations before increasing butanol concentration. Our results showed that e. coli typically grew well in the 0.8-1.2% (vol/vol) range. Occasionally, cells from higher butanol concentrations would survive, but would often be killed in the next experimental run. Therefore, above 1.2%, there was no consistent butanol tolerance.
