Virginia Tech
From 2007.igem.org
(Difference between revisions)
BlairLyons (Talk | contribs) m |
BlairLyons (Talk | contribs) m |
||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<html><center></html><!--first table: Title--> | <html><center></html><!--first table: Title--> | ||
| - | {| | + | {| style="border: 0px solid #FFCC66; width: 750px; color: #000000; background-color: #FFFFFF;" |
| Line 7: | Line 7: | ||
<center>[[Image:Title_bar.JPG]]</center> | <center>[[Image:Title_bar.JPG]]</center> | ||
| - | |||
| - | |||
|} | |} | ||
<!--second table: Navigation--> | <!--second table: Navigation--> | ||
| - | {| cellspacing="0px" border="0" style="width: | + | {| cellspacing="0px" border="0" style="width: 800px; color: #000000; background-color: #FFFFFF;" |
| Line 19: | Line 17: | ||
| style="background-color: #FFFFFF;" | | | style="background-color: #FFFFFF;" | | ||
| - | + | <html><a href="https://2007.igem.org/Virginia_Tech/Project"> | |
| + | <img src="https://static.igem.org/mediawiki/2007/0/04/Button_project.JPG"/></a></html>[[Image:Dot1.JPG]] | ||
<!--second cell of second table: Our Team--> | <!--second cell of second table: Our Team--> | ||
| style="padding: 0px; width=50px; background-color: #FFFFFF;" | | | style="padding: 0px; width=50px; background-color: #FFFFFF;" | | ||
| - | + | <html><a href="https://2007.igem.org/Virginia_Tech/Team"> | |
| + | <img src="https://static.igem.org/mediawiki/2007/2/2e/Button_team.JPG"/></a></html>[[Image:Dot1.JPG]] | ||
<!--third cell of second table: Progress--> | <!--third cell of second table: Progress--> | ||
| style="padding: 0px; width=50px; background-color: #FFFFFF;" | | | style="padding: 0px; width=50px; background-color: #FFFFFF;" | | ||
| - | + | <html><a href="https://2007.igem.org/Virginia_Tech/Updates"> | |
| + | <img src="https://static.igem.org/mediawiki/2007/3/3b/Button_progress.JPG"/></a></html>[[Image:Dot1.JPG]] | ||
<!--fourth cell of second table: Journal Club--> | <!--fourth cell of second table: Journal Club--> | ||
| style="padding: 0px; width=50px; background-color: #FFFFFF;" | | | style="padding: 0px; width=50px; background-color: #FFFFFF;" | | ||
| - | + | <html><a href="https://2007.igem.org/Virginia_Tech/JC"> | |
| + | <img src="https://static.igem.org/mediawiki/2007/d/d5/Button_jc.JPG"/></a></html>[[Image:Dot1.JPG]] | ||
| - | <!--fifth cell of second table: | + | <!--fifth cell of second table: Contributions--> |
| style="padding: 0px; width=50px; background-color: #FFFFFF;" | | | style="padding: 0px; width=50px; background-color: #FFFFFF;" | | ||
| - | + | <html><a href="https://2007.igem.org/Virginia_Tech/Contributions"> | |
| + | <img src="https://static.igem.org/mediawiki/2007/6/69/Button_contribute.JPG"/></a></html>[[Image:Dot1.JPG]] | ||
<!--sixth cell of second table: Contact--> | <!--sixth cell of second table: Contact--> | ||
| style="padding: 0px; width=50px; background-color: #FFFFFF;" | | | style="padding: 0px; width=50px; background-color: #FFFFFF;" | | ||
| - | + | <html><a href="mailto:igem@vt.edu"> | |
| + | <img src="https://static.igem.org/mediawiki/2007/d/da/Button_contact.JPG"/></a></html> | ||
|}<html></center></html> | |}<html></center></html> | ||
| - | <!--third table: | + | <!--third table: Content 1--> |
| - | {| cellspacing="0px" border="0" style="width: | + | {| cellspacing="0px" border="0" style="width: 880px; color: #000000; background-color: #FFFFFF;" |
<!--first cell of the third table: Space--> | <!--first cell of the third table: Space--> | ||
| + | | style="width: 0px; background-color: #FFFFFF" | | ||
| + | |||
| + | <!--second cell of the third table: Content 1--> | ||
| + | | style="padding: 20px; background-color: #FFFFFF;" | | ||
| + | |||
| + | '''<html><center><font size="5" color="#8b0000">The 2007 Virginia Tech iGEM team welcomes you to our Wiki! </font><br><br><font size="4">Look around to learn about our 2007 project: Engineering an Epidemic.</font></center></html>''' | ||
| + | |||
| + | |||
| + | |||
| + | <!--nested table: Content 2--> | ||
| + | {| cellspacing="0px" border="0" style="width: 880px; color: #000000; background-color: #FFFFFF;" | ||
| + | |||
| + | <!--first cell of the nestedtable: Space--> | ||
| style="width: 50px; background-color: #FFFFFF" | | | style="width: 50px; background-color: #FFFFFF" | | ||
| - | <!--second cell of the | + | <!--second cell of the nestedtable: left column--> |
| - | + | | style="padding: 20px; width: 400px; background-color: #FFFFFF" | | |
| - | | style="padding: | + | |
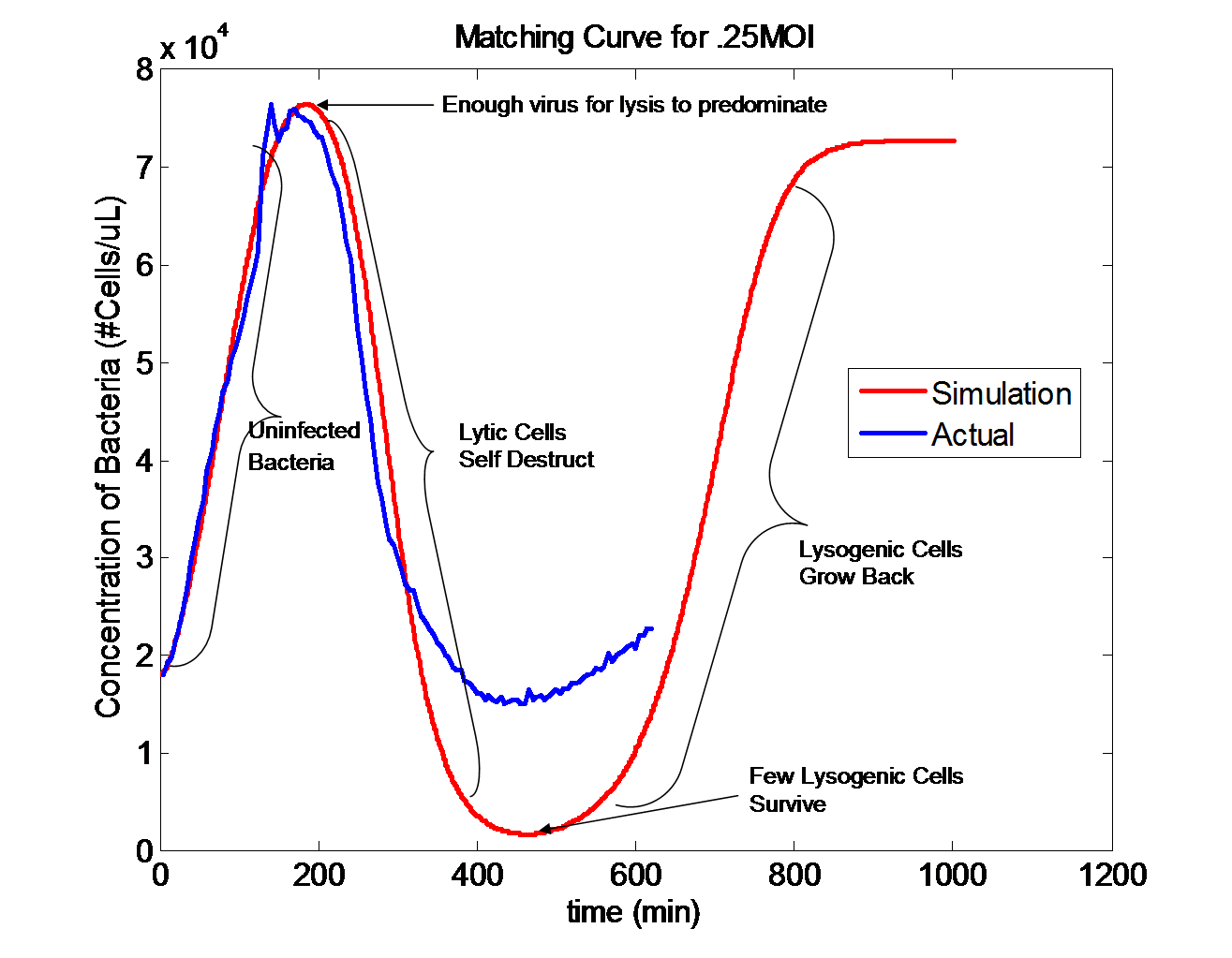
| - | + | [[Image:InfectionMatch.png|thumb|300px|center|'''Matching infection in a single population.''' Go to our | |
| + | [[Virginia_Tech/model|modeling page]] to learn more.]] | ||
| - | < | + | <html><center></html><h3>Modeling and Epidemiology</h3> |
| - | < | + | This year, our team worked on engineering an epidemic by simulating the spread of an infection in a biological model system. Our model has multiple levels, which we matched to experimental data one at a time. Click [[Virginia_Tech/model|here]] to read more about our modeling experience.<html></center></html> |
| - | < | + | |
| + | <!--third cell of the nested table: divider--> | ||
| + | | style="width: 5px; background-color: #FFFFFF;" | | ||
| + | |||
| + | <!--fourth cell of the nested table: right column--> | ||
| + | | style="padding: 20px; width: 400px; background-color: #FFFFFF;" | | ||
| + | |||
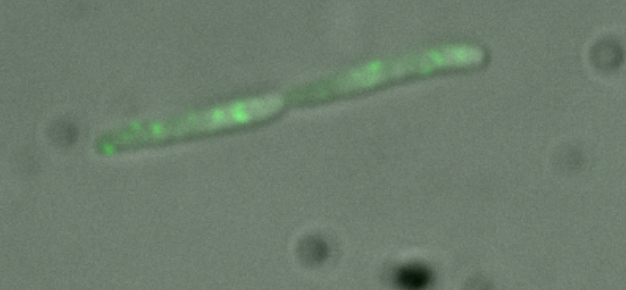
| + | [[Image:Ecoli phage.PNG|thumb|center|350px|'''''E.coli'' infected with fluorescent phage.''' This cell is lytic: the green spots are new phage particles being constructed prior to lysis.]] | ||
| + | |||
| + | <html><center></html><h3>Using a Biological Model System</h3> | ||
| + | λ phage and ''E. coli'' constitute our model system. λ phage has two infection pathways: lysis and lysogeny. A lytic cell will burst shortly after infection, while a lysogenic cell stays dormant. In order to find out which pathway had been chosen in our bacteria, we constructed a [[Virginia_Tech/plasmid_design|reporter plasmid]].<html></center></html> | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | <!--fourth table: Divider--> | ||
| + | {| cellspacing="0px" border="0" style="width: 880px; color: #000000; background-color: #FFFFFF;" | ||
| + | |||
| + | <!--first cell of the fourth table: Space--> | ||
| + | | style="padding: 5px; width: 50px; background-color: #FFCC66; border: 0px solid #000000;" | | ||
| + | |||
| + | |} | ||
| - | |||
| - | |||
| - | |||
| - | < | + | <!--fifth table: Sponsors--> |
| + | {| cellspacing="0px" border="0" style="width: 880px; color: #000000; background-color: #FFFFFF;" | ||
| - | < | + | <!--first cell of the fifth table: Space--> |
| - | + | | style="padding: 5px; width: 50px; background-color: #FFFFFF; border: 0px solid #000000;" | | |
| - | < | + | <!--second cell of the fifth table: Other Sponsors--> |
| - | + | | style="padding: 5px; background-color: #FFFFFF; border: 0px solid #000000;" | | |
| - | < | + | <h1>We would like to thank our sponsors:</h1> |
| - | < | + | <html><center></html><!--first nested table: first line--> |
| - | + | {| cellspacing="0px" border="0" style="color: #000000; background-color: #FFFFFF;" | |
| - | < | + | <!--second cell of the first nested table: DNA 2.0--> |
| + | | style="padding: 5px; background-color: #FFFFFF; border: 0px solid #000000;" | | ||
| - | < | + | <html><a href="http://www.dnatwopointo.com"> |
| - | <img src=" https://static.igem.org/mediawiki/2007/ | + | <img src="https://static.igem.org/mediawiki/2007/c/ce/DNA20_logo.jpg" width="150"></a></html> |
| - | < | + | <!--third cell of the first nested table: VBI--> |
| + | | style="padding: 5px; background-color: #FFFFFF; border: 0px solid #000000;" | | ||
| - | < | + | <html><center><a href="http://www.vbi.vt.edu"> |
| - | <img src="https://static.igem.org/mediawiki/2007/ | + | <img src="https://static.igem.org/mediawiki/2007/b/b4/Logo_VBI.JPG" width="150"></a></center></html> |
| - | <!-- | + | <!--fourth cell of the first nested table: VT--> |
| - | | style="padding: | + | | style="padding: 5px; background-color: #FFFFFF; border: 0px solid #000000;" | |
| - | + | ||
| - | + | <html><a href="http://www.vt.edu"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2007/b/b4/VTsponsor_logo.JPG" width="150"></a></html> | |
| - | + | |}<html><center></html> | |
| - | |||
| - | |||
| - | |||
| - | < | + | <!--third cell of the fifth table: Third Security--> |
| - | + | | style="padding: 5px; width: 0px; background-color: #FFFFFF; border: 0px solid #000000;" | | |
| - | + | ||
| + | <html><a href="http://www.thirdsecurity.com"><center><img src="https://static.igem.org/mediawiki/2007/a/a4/Sponsor_3rdsec2.JPG" width="150"> | ||
| + | <br></br><img src="https://static.igem.org/mediawiki/2007/6/65/Sponsor_3rdsec1.JPG" width="200"></center></a></html> | ||
|} | |} | ||
Latest revision as of 04:03, 26 October 2007
 |
|
Look around to learn about our 2007 project: Engineering an Epidemic.
|