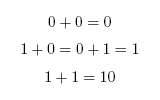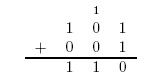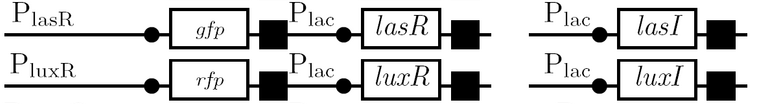Waterloo
From 2007.igem.org
(→Binary Addition) |
(→Binary Addition) |
||
| Line 11: | Line 11: | ||
===Binary Addition=== | ===Binary Addition=== | ||
| - | <div style="float:right;width: | + | <div style="float:right;width:200px;"> |
| - | <table cols="2" align=" | + | <table cols="2" width="150" align="left" style="border-collapse: collapse; padding-top:15px; padding-bottom:15px;"> |
<tr> | <tr> | ||
| - | <td colspan="1">Decimal<br>(Base 10) </td> | + | <td colspan="1" style="background-color:#CCCCCC; text-align:left;">Decimal<br>(Base 10) </td> |
| - | <td colspan="1">Binary <br>(Base 2)</td> | + | <td colspan="1" style="background-color:#CCCCCC; text-align:left;">Binary <br>(Base 2)</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td><div align="right">0</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">0</div></td> |
| - | <td><div align="right">00</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">00</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td><div align="right">1</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">1</div></td> |
| - | <td><div align="right">01</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">01</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td><div align="right">2</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">2</div></td> |
| - | <td><div align="right">10</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">10</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td><div align="right">3</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">3</div></td> |
| - | <td><div align="right">11</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">11</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td><div align="right">4</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">4</div></td> |
| - | <td><div align="right">100</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">100</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td><div align="right">5</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">5</div></td> |
| - | <td><div align="right">101</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">101</div></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td><div align="right">6</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">6</div></td> |
| - | <td><div align="right">110</div></td> | + | <td style="border-top: 1px solid black; border-bottom: 1px solid black;"><div align="right">110</div></td> |
</tr> | </tr> | ||
</table> | </table> | ||
Revision as of 08:20, 26 October 2007
 University of Waterloo iGEM Team
University of Waterloo iGEM Team Contents |
Our Team
The UW iGEM team is a very interdisciplinary group. Our team members span the three faculties of Science, Mathematics and Engineering and include the programs of Biology, Biomedical Sciences, Computer Science, Bioinformatics, Computer Engineering, Electrical Engineering, Chemical Engineering, and Mathematical Physics at undergraduate and graduate levels. Even our professor advisors are cross-appointed to two other faculties. Our diverse backgrounds bring together a wide range of skills and ideas to the iGEM project. iGEM is giving us the opportunity to apply the skills learned in our lectures and labs to real life applications in molecular biology and biotechnology.
Abstract
Background
Binary Addition
| Decimal (Base 10) |
Binary (Base 2) |
0 |
00 |
1 |
01 |
2 |
10 |
3 |
11 |
4 |
100 |
5 |
101 |
6 |
110 |
When working in binary, only two digits are used: 0 and 1. A single binary digit is called a bit. Counting proceeds as: To add two binary numbers, the process is much the same as adding two decimal (ordinary) numbers, except that instead of carrying when two digits add to ten, carrying must be performed when two digits add to two. In other words, 0 + 1 adds to 1, but 1 + 1 adds to 0 with a carry of 1, which gives 10 (just as in decimal 1 + 9 would add to 0 with a carry of 1, to give 10). A complete list of the possibilities is as follows:
Here is an example of long addition:
Half-Adder vs. Full-Adder
Any construct designed to add two numbers will either be a half-adder or a full-adder. A half-adder can only add together two single digits, whereas a full-adder is needed to add two numbers consisting of more than one digit.
For example, a half-adder could perform the addition 1 + 0 = 1, or 1 + 1 = 10, but it would take a full-adder to be able to perform 1100101 + 100101.
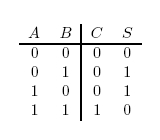
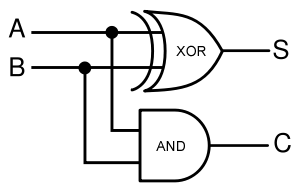
In terms of implementation, a half-adder accepts two inputs (the two digits to be summed) and returns two outputs (the "sum bit" and the "carry bit"). To add 1 + 1, the two inputs would each be 1, the sum bit would be 0, and the carry bit would be 1. A full list of the possibilities is shown in Figure 1.
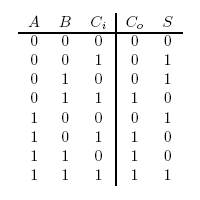
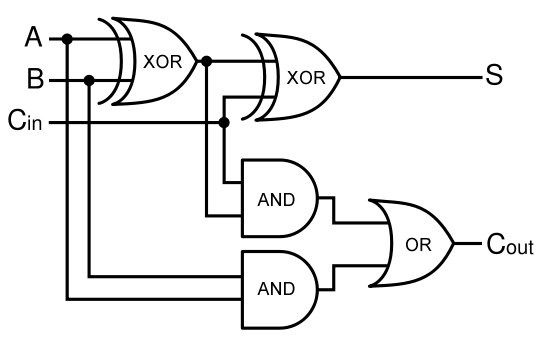
A full-adder is merely a half-adder that accepts an extra input; namely, the carry bit from another full-adder. Each full-adder is responsible for adding one pair of corresponding digits from the two numbers to be added, and it must add to that the carry bit from the previous full-adder. The full-adder will output the resulting sum bit and carry bit, and the process will continue until all the digits have been added. Such a chain of full-adders is called a ripple carry adder. See Figure 2 for a full list of possibilities.
Logic Gates and Implementing an Adder
A logic gate performs a logical operation on one or more logic inputs and produces a logic output. Usually the inputs and outputs are binary, meaning they each have a value of either 0 or 1. Also, it is often useful to think of of 0 as representing "false", and 1 as representing "true". A list of relevant logic gates is as follows:
- OR gate
- XOR gate
- AND gate
- NOT gate
For the following explanations, A and B represent statements that can be either true or false.
OR Gate
Consider the statement "A OR B". If either A or B is true, the statement is said to be true. The only time this statement will be false is when both A and B are false. This is the meaning of the OR operation.
An OR gate accepts two binary inputs, where a 1 represents "true" and a 0 represents "false". The output of the OR gate is also binary, and similarly represents either "true" or "false".
To illustrate,
0 OR 0 = 0
1 OR 0 = 0 OR 1 = 1
1 OR 1 = 1
XOR Gate
XOR is an abbreviation for "exclusive OR". The XOR gate is similar to the OR gate, the only difference being that it will return false if both inputs are true.
For example,
1 XOR 0 = 1
1 XOR 1 = 0
AND gate
The AND gate will return 1 (true) only if both the inputs are 1 (true). Otherwise, it will return a 0.
To illustrate,
0 AND 0 = 0
1 AND 0 = 0 AND 1 = 0
1 AND 1 = 1
NOT gate
The NOT gate accepts one input, and simply switches its value.
NOT 1 = 0
NOT 0 = 1
Using logic gates to make an adder
Given two binary inputs A and B, their sum can be computed by passing these inputs through various logic gates. The process is illustrated in Figures 4 and 5.
DNA as Logic Gates
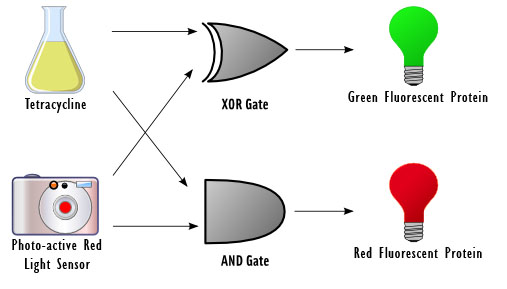
The focus of our project revolves around using DNA as logic gates. In essence, the output of a logical operation is the chemical produced by a certain gene, and the inputs are external stimuli that modify the conformation of proteins, which act on the promoters of the genes to induce or inhibit transcription depending on the specific logic gate implemented.
For example, consider a positively regulated gene G that codes for protein X. By default, the gene is not expressed. If the inputs A and B are on and inducing protein activity that turns on G, the expressed protein represents the output of an AND gate. The inputs would be the "presence" or "absence" of A and the "presence" or "absence" of B (both representing 0 and/or 1 as inputs), and the output would be the "presence" or "absence" of X (where "presence" and "absence" should be read as high and low intensity/concentration, respectively).
This is the type of approach used to construct our biological adder. Further details are given under the Project Details section.
Project Details
Inputs
As a half-adder, our cells require two binary inputs. We chose to use the presence or absence of red light as one input, and the presence or absence of the chemical tetracycline as another. For the first input, the presence of red light represents a 0 and the absence represents a 1. For the second input, the absence of tetracycline represents a 0 and the presence represents a 1.
We chose to use light as an input for the reasons that it does not require altering the media, it can be turned on and off rapidly, and it has extremely high resolution and accuracy. The red light will be detected by the previously used Cph1/EnvZ fusion protein (BBa_I15010).
Tetracycline was used as a second input since it binds to the tetR repressor ultimately allowing for the production of the Green Fluorescent Protein (GFP) or Red Fluorescent Protein (RFP). It also will not interfere with the constitutive promoter and can be easily controlled.
Output
Depending on the result of the calculation, the bacteria will produce either red fluorescent protein (RFP), green fluorescent protein (GFP), or neither. The possible outcomes are as follows:
No Tetracycline & No red light ==> 0+0 ==> nothing
No Tetracycline & Red light ==> 0+1 ==> GFP
Tetracycline & No red light 1+0 ==> GFP
Tetracycline & red light ==> 1+1 ==> RFP
A diagram of the biological half adder in culture is shown below. It shows all outputs given all possible inputs.
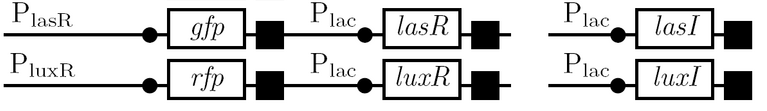
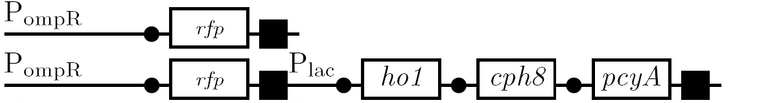
Schematic Diagram
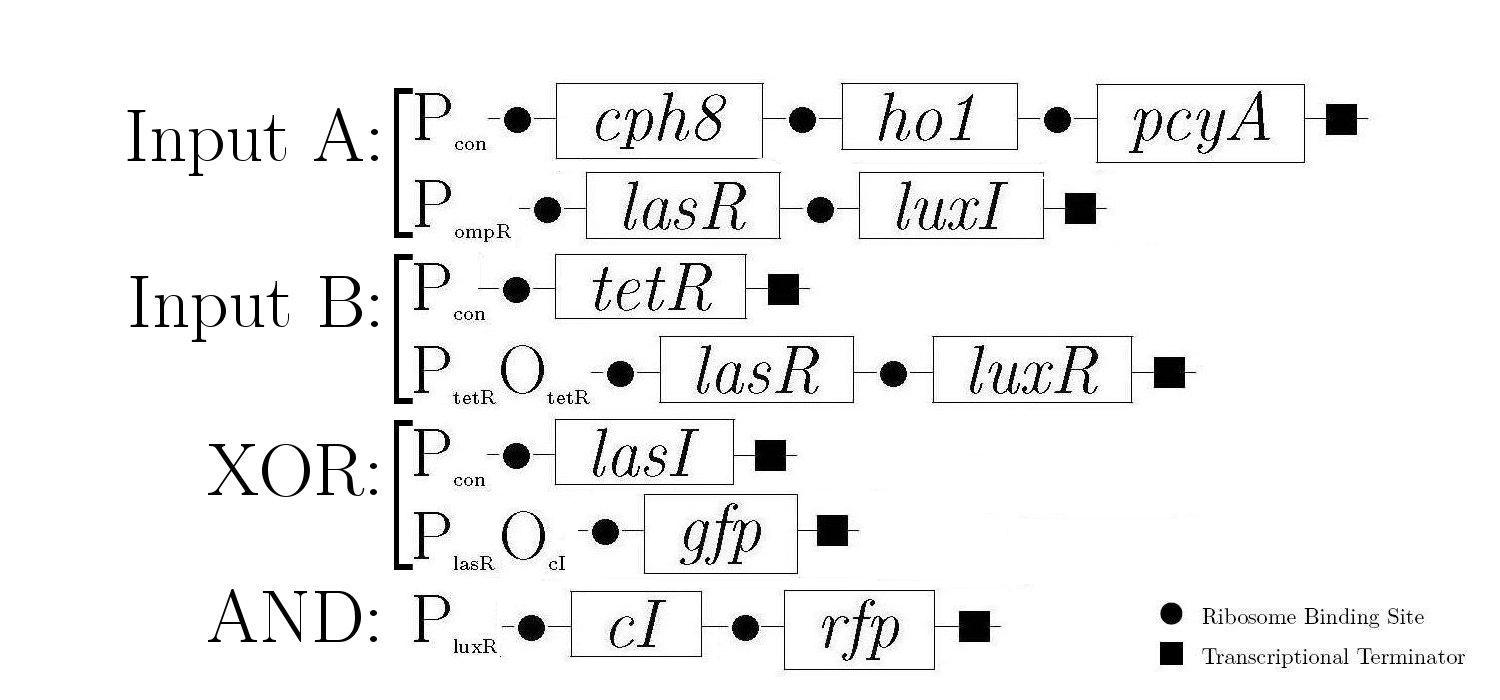
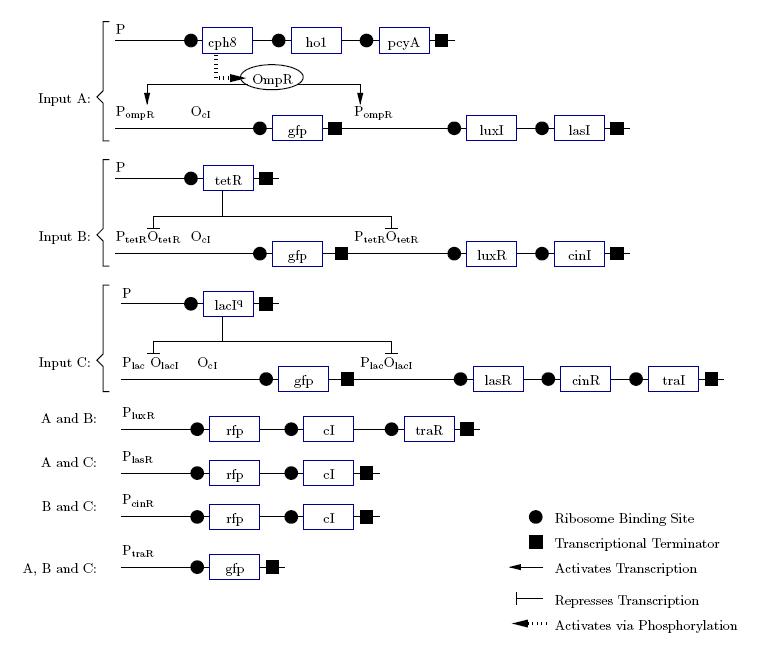
Gene Diagram
Construction
Testing/Results
Testing Constructs
Making use of flurorescent proteins(GFP and RFP) as reporter genes - use of indicuble promoters to...
Comparing basal and induced expression levels for the quorum sensing promoters
Testing functionality of the various promoters
Mathematical Model
To create a predictive model, we used a set of differential equations to graph the approximate concentrations of each chemical with respect to time. These graphs are intended to illustrate the relative concentrations of the chemicals involved for each possible combination of inputs. The first quarter of the graph (t=0 to 500) illustrates the concentration of chemicals when adding 0+0; the second quarter (t=500 to 1000) illustrates the 0+1 case; the third quarter illustrates the 1+0 case; and the fourth quarter the 1+1 case.
<math>\frac{d[O]}{dt} = k' + \frac{k \cdot ([I]^n/K^n)^\mu}{1 + ([I]^n/K^n)^\mu} - d \cdot [O] </math>
Measurements
Extensions
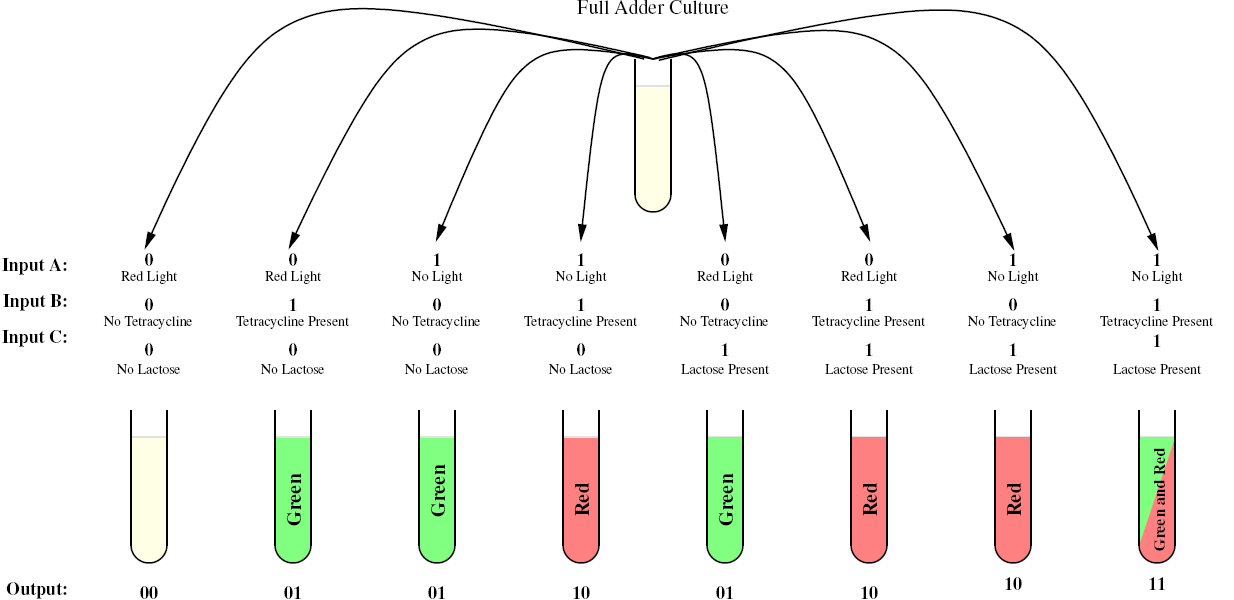
Full Adder
As a future project, we would like to extend our half-adder to a full-adder, which would be much more useful as it would give us the ability to add numbers of more than one digit. Below is a diagram of all the outputs given all possible inputs.
A gene design that makes use of some of the half-adder components can be seen below.
Acknowledgements
The UW-iGEM team would like to thank the following organizations and individuals for their contributions:
Faculty of Science Foundation of University of Waterloo |
Mathematics Endowment Fund of University of Waterloo |
Sir Sanford Fleming Foundation of University of Waterloo |
|
- Dr. Trevor Charles