Edinburgh/DivisionPopper
From 2007.igem.org
(→Assumptions) |
(→Exp 2) |
||
| Line 40: | Line 40: | ||
[[Image:Edinburgh_Divisiontest2.png|300px|float|left]] | [[Image:Edinburgh_Divisiontest2.png|300px|float|left]] | ||
| - | This | + | This investigates the number of flips that occurs during division and will require rapidly degrading fluorescent proteins. |
| - | After each division we expect to see a change in colour as the promoter | + | After each division we expect to see a change in colour as the promoter activates a different reporter. |
====Exp 3==== | ====Exp 3==== | ||
Revision as of 12:15, 23 July 2007
https://static.igem.org/mediawiki/2007/f/f5/800px-Edinburgh_City_15_mod.JPG
POPS Oscillator reporting cell division
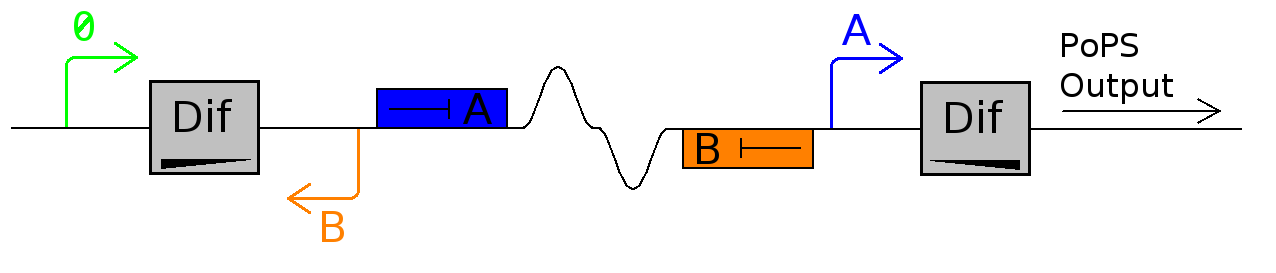
The idea is to use the XerC and D recombinases to invert a section of DNA at cell division. Inversion of the DNA causes a PoPS pulse as output.
Project Goal
The aim is to produce output as a function of bacterial cell division. Different versions of the recombination enable different downstream outputs. The system may for example perform programmed cell death after a set number of cell divisions. We hope to further analyse cell division and recombinase mechanisms since bacterial cell division is still relatively poorly understood.
Current Device Idea
This device outputs a PoPS pulse at each cell division. It can then be hooked up to another device such as a counter.
For this device to work, dif site-enclosed DNA needs to flip but once per division. Like all recombination sites dif-sites are directional and bacteria uses directly repeated dif-sites to resolve genome dimers. Research shows that this resolution occurs only at septation, and we use this temporal control to inverse a plasmid-kept sequence to induce differential functions. In it's current configuration, Promoter A is repressed by continually produced represser A. At this time, no represser B is produced.
As cell division occurs, DNA enclosed by two inverted dif sites is reversed. Initially there is no represser B present in the cell so promoter B produces PoPS output. As time passes, represser B is produced and 'turns off' promoter B. Meanwhile represser A is no longer produced and degrades so that, at the next division, promoter A can produce PoPS output and the process repeats.
Assumptions
- The Dif sites flip only once during cell division
- Repressors A and B are produced and degrade faster than the cell cycle
- Promoter B does not interfere with production of repressor A
Initial Experiments
As stated before, the device relies on the dif site flipping only once per cell division. To test whether or not this actually happens, we have devised two simpler experiments.
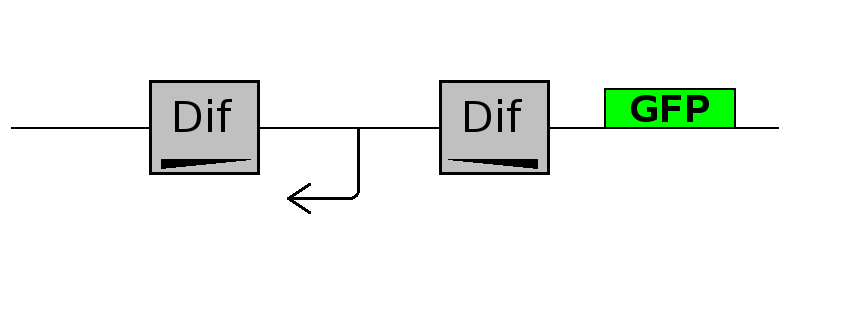
Exp 1
This experiment will prove whether or not the DNA between the two dif sites flip at all during cell division. If the a flip occurs, then the direction the promoter operates will be changed and GFP will be expressed.
Exp 2
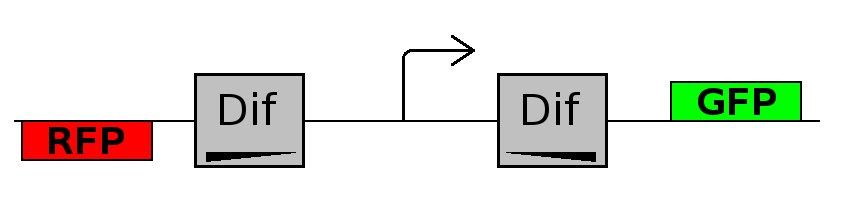
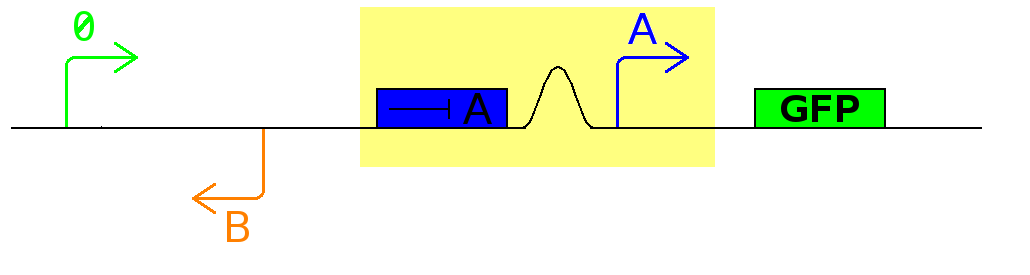
This investigates the number of flips that occurs during division and will require rapidly degrading fluorescent proteins.
After each division we expect to see a change in colour as the promoter activates a different reporter.
Exp 3
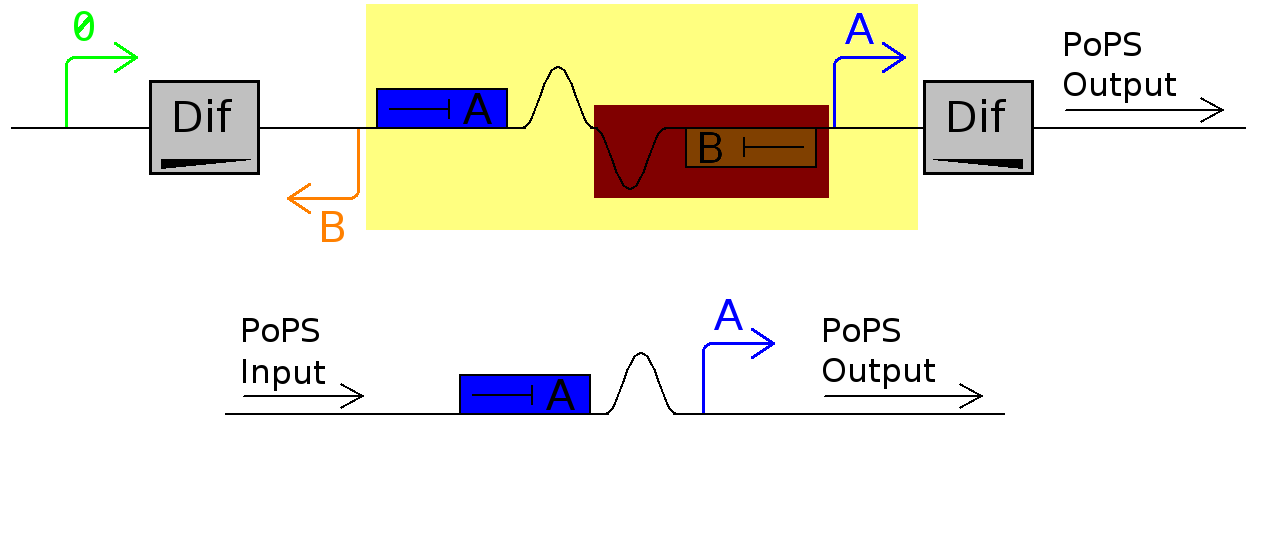
Currently we are planning to test how the dif sites flip using exps 1 & 2 however there is still a large jump to the final device.
It was suggested we should test the represser part of the system without the dif sites
The represser part of the system is simply a pair of inverters.
There are 3 basic types of inverter in the registry:
- TetR BBa_Q04400 “this inverter functions well. [jb, 5/24/04]”
- CI (Lambda) BBa_Q04510 “this inverter functions well. [jb, 5/24/04]”
- LacI BBa_Q04121 "a strong 'on' state with significant background in the 'off' state. [jb,5/24/04]”
Suggested exp 3
This will test to see if promoter B causes problems with promoter 0 promoting represer A and tests the standard represser inverter found in the registry.
Parts required:
- Promoter 0 – suggest BBa_I0500 (arabinose) – Plate 2, 9I
- Promoter B – backward lacI – Used in exp 1
- Inverter – suggest BBa_Q04510 (CI (Lambda)) – Plate 2, 13K
- Reporter – use same as experiment 1
References
[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1449051&blobtype=pdf Bloor - An Efficient Method of Selectable Marker Gene Excision by Xer Recombination for Gene Replacement in Bacterial Chromosomes (2006)]
- This article describes a system where a gene is deleted by replacing it with an antibiotic marker. The antibiotic marker is then excised during next cell divsion using the naturally expressed XerCD? recombinases. This is similar to what we are doing except instead of excising we are inverting.
| "The natural dif site is in the chromosome terminus region and is essential for correct chromosome segregation at cell division: deletion of difE. coli results in a subpopulation of E. coli cells with a filamentous morphology (12). However, if natural difE. coli is deleted, another difE. coli site will not enable dimer resolution if placed outside of the 30-kb dif activity zone (DAZ) in the terminus region (6, 16). Therefore, while our method involves insertion of one or more extra dif sites at different chromosomal loci, this should not have an adverse effect on chromosome segregation, although the possibility that introduction of multiple dif sites in close proximity might result in deletion of intervening chromosomal DNA should be considered." |
- Summary: Dif sites outside of the DAZ zone shouldn't cause problems with the natural dif sites of the chromosome/plasmid
[http://www.blackwell-synergy.com/doi/full/10.1046/j.1365-2958.1998.00651.x Kuempel - Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli (1998)]
- Good overview of the natural Xer recombinase system.