Paris/August 7
From 2007.igem.org
(Difference between revisions)
(→Study of the pBAD promoter) |
(→Study of the pBAD promoter) |
||
| Line 1: | Line 1: | ||
== Study of the pBAD promoter == | == Study of the pBAD promoter == | ||
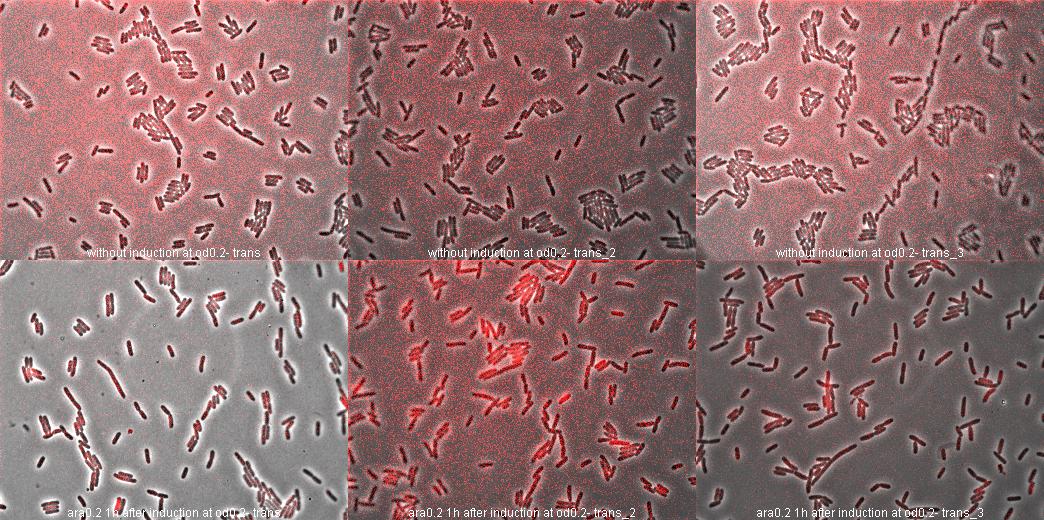
We've put the AraC-pBAD promoter (<bbpart>BBa_I0500</bbpart>) in the expression vector <bbpart>BBa_J61002</bbpart> (between EcoRI and SpeI). After overnight culture of E.coli transformed by this plamid, we diluted 1/100 and waited 2 hours. At OD=0.2, we divided the culture in 2 subcultures, one with arabinose (final 0.2%) and one without. | We've put the AraC-pBAD promoter (<bbpart>BBa_I0500</bbpart>) in the expression vector <bbpart>BBa_J61002</bbpart> (between EcoRI and SpeI). After overnight culture of E.coli transformed by this plamid, we diluted 1/100 and waited 2 hours. At OD=0.2, we divided the culture in 2 subcultures, one with arabinose (final 0.2%) and one without. | ||
| - | |||
* '''1 hour''' later, we took pictures with fluorescence microscope (triplicate): <br> | * '''1 hour''' later, we took pictures with fluorescence microscope (triplicate): <br> | ||
| Line 7: | Line 6: | ||
- the second represent subculture with 0.2% arabinose induction. | - the second represent subculture with 0.2% arabinose induction. | ||
| - | [[Image: t0_pBad-AraC.jpg|center|600px]] | + | [[Image: t0_pBad-AraC.jpg|center|600px]]<br> |
| - | + | ||
| - | <br> | + | |
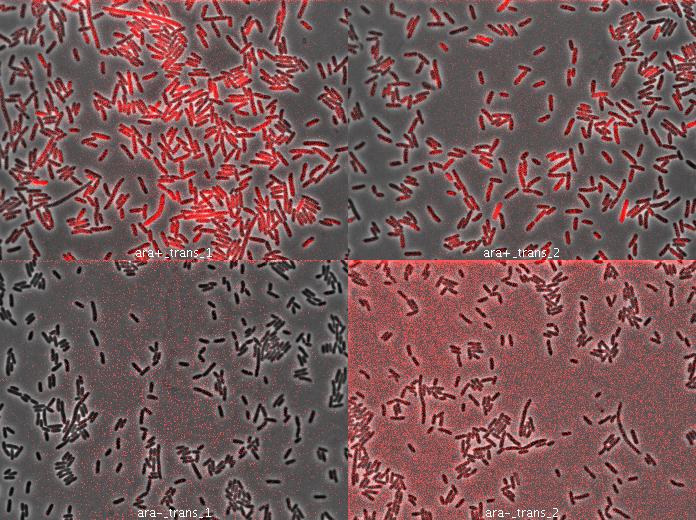
* '''5 hours''' later: | * '''5 hours''' later: | ||
[[Image: t5_pBad-AraC.jpg|center|600px]] | [[Image: t5_pBad-AraC.jpg|center|600px]] | ||
| + | |||
| + | * '''Interpretation:''' | ||
| + | |||
| + | Arabinose induction looks to work. mRFP expression looks to increase with time (the acquisition and analyse were identical at each time). | ||
== Digestion reactions == | == Digestion reactions == | ||
Revision as of 08:51, 8 August 2007
Study of the pBAD promoter
We've put the AraC-pBAD promoter (BBa_I0500) in the expression vector BBa_J61002 (between EcoRI and SpeI). After overnight culture of E.coli transformed by this plamid, we diluted 1/100 and waited 2 hours. At OD=0.2, we divided the culture in 2 subcultures, one with arabinose (final 0.2%) and one without.
- 1 hour later, we took pictures with fluorescence microscope (triplicate):
- the first line represent subculture without arabinose induction. - the second represent subculture with 0.2% arabinose induction.
- 5 hours later:
- Interpretation:
Arabinose induction looks to work. mRFP expression looks to increase with time (the acquisition and analyse were identical at each time).
Digestion reactions
| Digestion Products | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Product Name | Matrix Name | Enzyme 1 | Enzyme 2 | Size | Description | |||
| D37'.1 | lox71-gfp(tripart) BI | L12.1 miniprep product (from 30/07 ligation reaction) | XbaI | PstI | lox71_RBS-rfp-double terminator extracted as a backward insert | ||||
| D39' | BI-insert lox66-DapAE.coli | lox66-DapAE.coli in pTOPO (P5 PCR product)(clones 1+2) | XbaI | PstI | lox66-dapAcoli extracted for usage as a backward insert | ||||
| D41 | araC/pBad-J61002 plasmid SP dig. | L1.2, L1".1 & L1".2 miniprep product | SpeI | PstI | J61002 plasmid containing araC/pBad promotor digested for insertion of a BI CDS | ||||
Minipreps
| Number | Name | Description |
| MPT2 | S16.2 | |
| MPT3.1 | pTOPO P6.1 | |
| MPT3.2 | pTOPO P6.7 | |
| MPT4.1 | pTOPO P8.2 | |
| MPT4.2 | pTOPO P8.5 | |
| MPT4.3 | pTOPO P8.7 | |
| MPT4.4 | pTOPO P8.8 | |
| MPT4.5 | pTOPO P8.10 | |
| MPT5.1 | pTOPO P9.1 | |
| MPT5.2 | pTOPO P9.2 | |
| MPT5.3 | pTOPO P9.4 |