Paris/What's going on in the lab
From 2007.igem.org
Nicolas C. (Talk | contribs) (→In progress) |
Nicolas C. (Talk | contribs) (→In progress : molecular biology part) |
||
| Line 1: | Line 1: | ||
See the [[Paris/Notebook Calendar|Notebook]] | See the [[Paris/Notebook Calendar|Notebook]] | ||
== In progress : molecular biology part == | == In progress : molecular biology part == | ||
| + | Red : done | ||
| + | Blue : fail | ||
| + | Black : in progress... | ||
Big construct to be inserted in the chromosome | Big construct to be inserted in the chromosome | ||
[[Image:Paris_Montage1.png]] | [[Image:Paris_Montage1.png]] | ||
Revision as of 16:40, 14 August 2007
See the Notebook
Contents |
In progress : molecular biology part
Red : done
Blue : fail
Black : in progress...
Big construct to be inserted in the chromosome
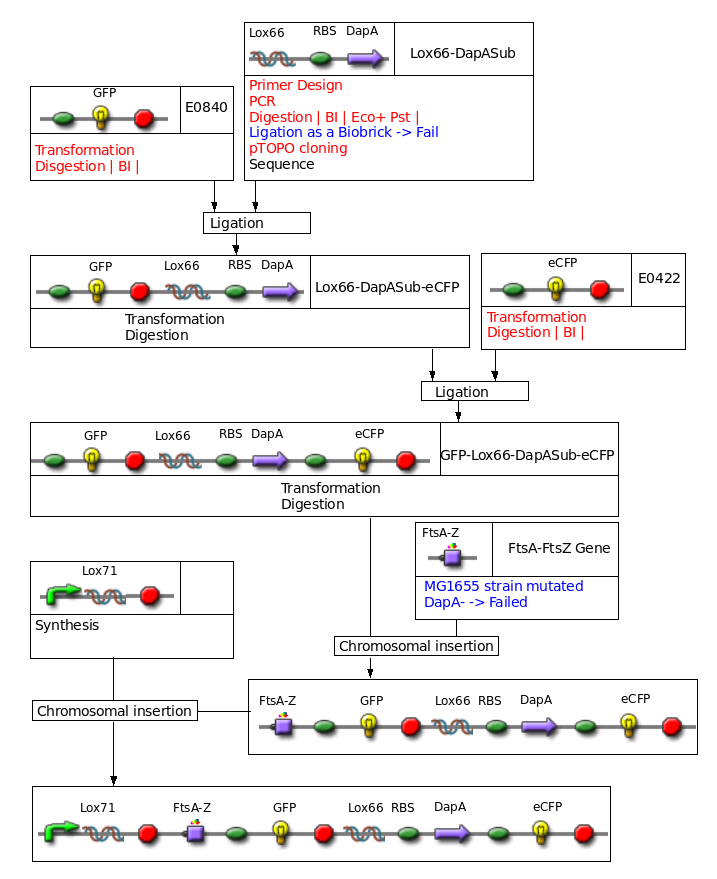 Construct : inducible CRE
Construct : inducible CRE
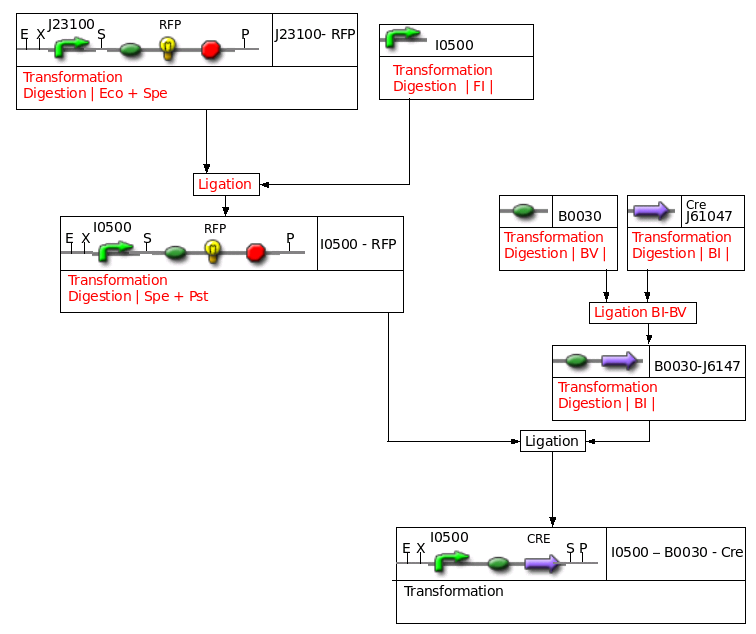 Miscellaneous
File:Paris Montagereste.png
Miscellaneous
File:Paris Montagereste.png
DGAT and some other things are missing
What's new ?
- Since we had dome trouble cloning our PCR products, we decided to clone them in the pTopo plasmid of invitrogen. This way we were able to clone the DapA gene of Coli and subtilis, but sill did not succeed in cloning FtsZ or FtsA+FtsZ... The insert in the pTopo is expressed, and our inserts are certainly toxic...
To Do List
- Cloning of Lox71-FtsZ and Lox71-FtsA-FtsZ in J61002 with the constitutive promoter and with the pBad
This will allow us to see the effect of the overexpression of FtsZ and FtsA+FtsZ on the growth and phenotype. We expect the overexpression of FtsZ only to be lethal and the one of FtsA+FtsZ to form a viable mini-cell phenotype.
- Cloning of Lox66-DapA of Coli and Subtilis in the same 2 vectors
In Progress
FstZ strain caracterisation
We try to obtain a reliable TS clone, which would not grow at 42°C, but grow at 30°C
Transduction of the deletion of DapA from the strain w121 into MG1655 and MG1655 FtsZ TS84
The goal is, by transduction, to introduce DapA deletion into FtsZ TS strain
- We managed to build a stock of efficient phages (stock of 12.7.7)
- Transduction in progress
Caracterisation of w121 - MG1655 feeding supplementation
This part is just a preliminary work whose aim is to show that two strains can complement each other. A more detailed explanation of these experiments can be found in the team notebook.
- Several conditions were tested but the curves are difficult to interpret. See preliminary results.
Caracterisation of Acinetobacter strain
Goal : in the final construct, we will take the gene DGAT from this strain and put it in E. Coli.
Preliminary work has been done on acinetobacter :
- We confirmed the production of TG in acinetobacter when it is grown on minimal medium + a source of carbon (ask David from precise information). We were able to stain TG with Nile Red fluorescent dye (cf photos):
- in a whole colony
- on single cells
- We began cloning of DGAT gene on E. Coli (ask David for precise information)
Accomplishments
- PCR and Cloning of lox71-KmR-Lox66
- Cloning of lox71 and lox66
- Cloning of I0500 (inducible pBad) in [http://partsregistry.org/Part:BBa_J61002 J61002]
This will allow us to compare the strength of the pBad and the strengh of the constitutive promoter. Moreover, we'll be able to clone any gene we like after the promoter.
Transformation of 8 Biobricks
The goal is to extract DNA from the plates and make a stock of plasmid we need.
- BBa_I0500: Inducible pBad/araC in pSB2K3 (KanR)
- BBa_J23100: strong constitutive promoter in BBa_J61002 (AmpR)
- BBa_B0015: double terminator (B0010-B0012) in pSB1AK3 (AmpR)
- BBa_B0030: RBS (well 3G plate 1)
- BBa_E0422: ECFP (RBS+LVA+Term) (well 11G plate 1)
- BBa_E0241: PoPs to GFP converter (well 15c plate 2)
- BBa_E0840: gfp tri-part; strong rbs (well 16E plate 1)
- BBa_J61047: Cre ORF (well 8P plate 4)