UCSF/Intro
From 2007.igem.org
< UCSF(Difference between revisions)
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | [[Image: | + | {| |
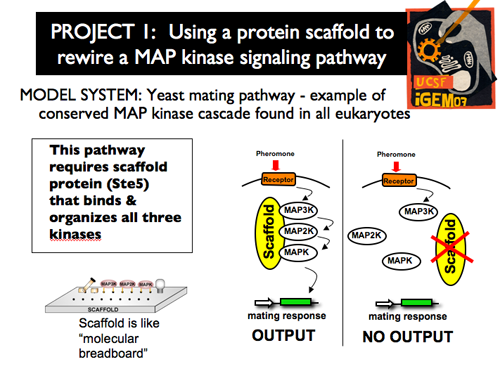
| - | [[Image: | + | |[[Image:p1intro9.png]]||'''In our first project, we treated the scaffold protein as a molecular breadboard. We chose to specifically work with the scaffold protein Ste5, which organizes the MAP kinases in the yeast mating pathway. Because this kinase cascade is conserved in all eukaryotes, it is closely examined and well-characterized. Responding to pheromone outside of the cell, yeast transduce an external cue through this cascade to ultimately reach the nucleus, where transcriptional changes cause a mating response. Ste5 is crucial in transmitting this signal—without the organizational help of the scaffold, the MAP3K, MAP2K and MAPK are unable to effectively communicate. As a result, the mating response cannot be produced.''' |
| - | [[Image: | + | |- |
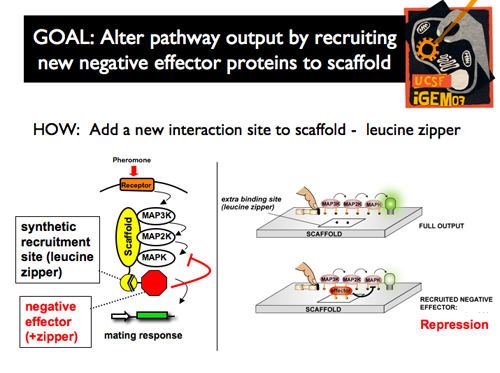
| - | [[Image: | + | |[[Image:p1intro10.png]]|| '''In our project, we plugged in an additional interaction site to the scaffold by fusing a synthetic leucine zipper domain to Ste5. This serves as an additional docking site for any protein that contains the complimentary half of the leucine zipper. Using this strategy, we recruited negative effectors that would repress the activity of the kinase cascade, and thus, modify the pathway output. We were able to measure the output of the pathway with a mating pathway induced GFP reporter.''' |
| - | + | |- | |
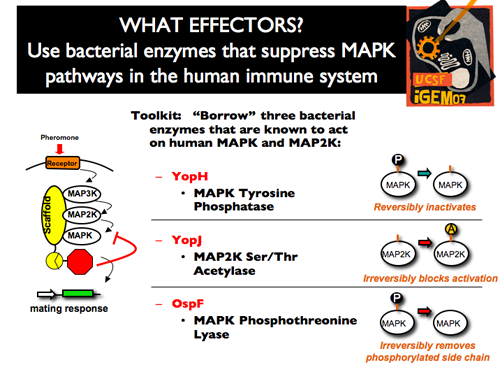
| - | + | |[[Image:p1intro11.png]]||'''Since we wanted to be able to effect a wide variety of dynamic changes, we created a toolkit of three different negative effectors, all of which were borrowed from bacteria. YopH reversibly inhibits the MAPK by cleaving the phosphate group on the tyrosine residue that determines the MAPK activity. YopJ, on the other hand, works irreversibly on the MAPKK by acetylation. Our third effector, OspF, is also an irreversible inhibitor, but it cleaves both the phosphate and the R-group from the MAPK regulation site. Because these three effectors vary in substrate, catalytic activity, and dynamics of inhibition, they form a very useful toolbox for engineering the mating pathway.''' | |
| - | + | |- | |
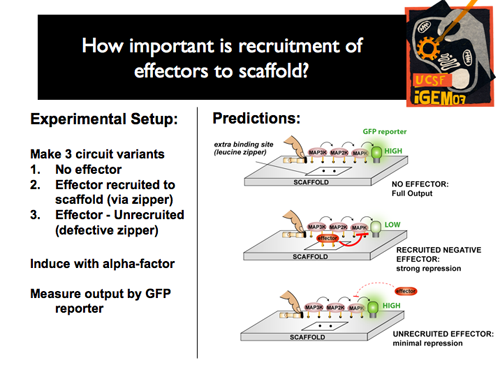
| - | [ | + | |[[Image:p1intro12.png]]||'''Experimentally, we wanted not only to compare the range of effects through the effector toolbox, but also to examine the significance of recruitment of each effector the scaffold. So, for each experiment, we created three variants: one with no effector, one with the effector recruited to the scaffold, and one with an effector that was unable to be recruited to the scaffold. We hypothesized that, compared to the full output of the undisturbed pathway, we should see a strong repression with the recruited effector, but only minimal repression without actual recruitment to the scaffold.''' |
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div style="text-align: right;"> <font size=4><font color=firebrick>Move on to see our '''[https://2007.igem.org/UCSF/Results Results!]''' </font></font></div> | ||
Latest revision as of 23:51, 28 September 2007
Move on to see our Results!