NYMU Taipei/Lab Notes/2007 10 4
From 2007.igem.org
< NYMU Taipei/Lab Notes(Difference between revisions)
(→Re-check the concentration of inserts and vectors) |
(→transformation setup) |
||
| (16 intermediate revisions not shown) | |||
| Line 27: | Line 27: | ||
</td> | </td> | ||
<td> | <td> | ||
| - | * | + | * Owing to |
| + | ** concentrations of vectors are too low and | ||
| + | ** A260/A280 ratios are not correct (maybe too much ions) | ||
| + | * thus, we checked the vectors with inserts by PCR to decide how to ligate them | ||
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | == ligation setup == | ||
| + | * criteria | ||
| + | ** make vector have around 100 ng in sample (total volume 20 uL) | ||
| + | ** maximize the insert volume, and insert is larger than vector | ||
| + | <table border=1 align=center> | ||
| + | <tr align=center> | ||
| + | <td>ligation I: pCinRHSL (vector) + OmpRBS (insert)</td> | ||
| + | <td>ligation II: pOmpC (vector) + TATA_INSA (insert)</td> | ||
| + | <td>ligation III: CinR+HSL+D-term (vector) + TATB_INSB (insert)</td> | ||
| + | </tr> | ||
| + | |||
| + | <tr align=center><td> | ||
| + | <table border=1 width=100%> | ||
| + | <tr><td>pCinRHSL (vector, 20 ng/uL)</td><td>3 uL (60 ng)</td></tr> | ||
| + | <tr><td>OmpRBS (insert, 30 ng/uL)</td><td>14 uL (420 ng)</td></tr> | ||
| + | <tr><td>10X buffer</td><td>2 uL</td></tr> | ||
| + | <tr><td>T4 ligase</td><td>1 uL</td></tr> | ||
| + | <tr><td>insert:vector</td><td>1:7</td></tr> | ||
| + | </table> | ||
| + | </td> | ||
| + | |||
| + | <td> | ||
| + | <table border=1 width=100%> | ||
| + | <tr><td>pOmpC (vector, 10 ng/uL)</td><td>4.5 uL (45 ng)</td></tr> | ||
| + | <tr><td>TATA_INSA (insert, 20 ng/uL)</td><td>12.5 uL (250 ng)</td></tr> | ||
| + | <tr><td>10X buffer</td><td>2 uL</td></tr> | ||
| + | <tr><td>T4 ligase</td><td>1 uL</td></tr> | ||
| + | <tr><td>insert:vector</td><td>1:5.5</td></tr> | ||
| + | </table> | ||
| + | </td> | ||
| + | |||
| + | <td> | ||
| + | <table border=1 width=100%> | ||
| + | <tr><td>CinR+HSL+D-term (vector, 20 ng/uL)</td><td>3 uL (60 ng)</td></tr> | ||
| + | <tr><td>TATB_INSB (insert, 30 ng/uL)</td><td>14 uL (420 ng)</td></tr> | ||
| + | <tr><td>10X buffer</td><td>2 uL</td></tr> | ||
| + | <tr><td>T4 ligase</td><td>1 uL</td></tr> | ||
| + | <tr><td>insert:vector</td><td>1:7</td></tr> | ||
| + | </table> | ||
| + | </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | |||
| + | == transformation setup == | ||
| + | * extract 4 uL from ligation mixture (total 20 uL) | ||
| + | * use whole competent cell (total 1 mL) for each ligation | ||
| + | * add ligation mixture into competent cell before it entirely melts and mix well | ||
| + | * 42 C for 45 sec. and put it into iced water (4 C) | ||
| + | * finish within around 5 mins (don't put the competent cell in iced water more than 5 mins) | ||
| + | |||
| + | == Next tasks == | ||
| + | * check the ligation by PCR | ||
| + | ** if the size is correct, perform plasmid extraction | ||
| + | ** else, ligate them again | ||
Latest revision as of 15:13, 4 October 2007
Contents |
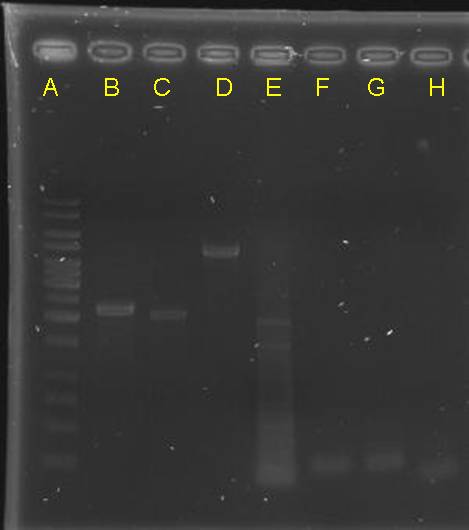
digestion check of CinR+HSL+D-term,pCinRHSL and pOmpC (vectors)
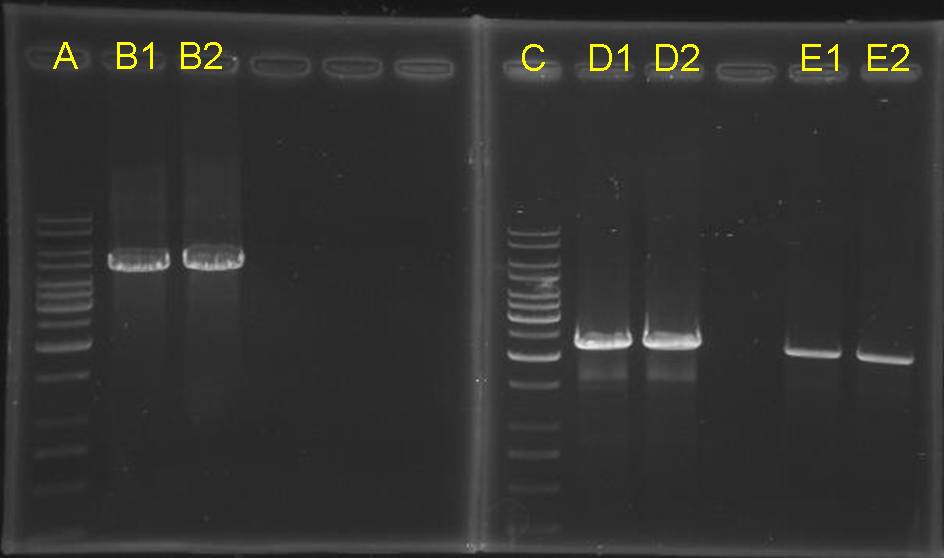 |
|
- after gel separation, their O.D. was checked. However, it is too low
Re-check the concentration of inserts and vectors
ligation setup
- criteria
- make vector have around 100 ng in sample (total volume 20 uL)
- maximize the insert volume, and insert is larger than vector
| ligation I: pCinRHSL (vector) + OmpRBS (insert) | ligation II: pOmpC (vector) + TATA_INSA (insert) | ligation III: CinR+HSL+D-term (vector) + TATB_INSB (insert) | ||||||||||||||||||||||||||||||
|
|
|
transformation setup
- extract 4 uL from ligation mixture (total 20 uL)
- use whole competent cell (total 1 mL) for each ligation
- add ligation mixture into competent cell before it entirely melts and mix well
- 42 C for 45 sec. and put it into iced water (4 C)
- finish within around 5 mins (don't put the competent cell in iced water more than 5 mins)
Next tasks
- check the ligation by PCR
- if the size is correct, perform plasmid extraction
- else, ligate them again