Paris/August 3
From 2007.igem.org
(→DGAT biobrick: digestion, cloning) |
Nicolas C. (Talk | contribs) |
||
| (27 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == DGAT biobrick: | + | [[Paris/August 2|yesterday]] -- [[Paris/August 4|tomorrow]] <br> |
| - | + | == DGAT biobrick: EcoR1-Pst1 digestion == | |
| - | + | ||
[[Image: DGATbbdigEcoR1Pst108032007.jpg|100px|right]] | [[Image: DGATbbdigEcoR1Pst108032007.jpg|100px|right]] | ||
| Line 7: | Line 6: | ||
The double digestion allows us to insert the biobrick into biobrick plasmid.<br> | The double digestion allows us to insert the biobrick into biobrick plasmid.<br> | ||
The Pst1 digestion allows us to see if the mutagenesis worked (dgat gene not digested). | The Pst1 digestion allows us to see if the mutagenesis worked (dgat gene not digested). | ||
| + | |||
| + | *'''Material:''' | ||
| + | - DNA (PCR product DGAT deltaPst-1 biobrick from [[Paris/August_2|August 2]]): 200ng = 20µL <br> | ||
| + | - BSA: 0.5µL<br> | ||
| + | - NEBuffer EcoR1: 5µL<br> | ||
| + | - EcoR1 enzyme: 1µL<br> | ||
| + | - Pst-1 enzyme: 1µL<br> | ||
| + | - ddH<sub>2</sub>O<br> | ||
| + | |||
| + | * '''Protocol:''' | ||
| + | - 2 hours at 37°C | ||
| + | |||
| + | * '''Purification''' | ||
| + | |||
| + | == Dap complementation experiment == | ||
| + | |||
| + | We used wild type E.coli transformed by part J23107 (RFP) and dapA- E.coli (w121) auxotroph to DAP. E.coli transformed by part J23107 (RFP) is able to synthesize DAP and export it. We suppose dap- E.coli could be feed by wt E.coli. | ||
| + | |||
| + | We tried to observe kinetics by fluorescence microscopy. We let separately in culture overnight dapA- E.coli (LB/erythromycine/DAP) and rfp wt E.coli (LB ampicillin). The day after, we diluted both liquid culture 1/200 to have exponential culture. At OD = 0.7 (dapA-) and OD = 0.5 (wt rfp), we mix both population in different condition culture (We used preliminary results ([[Paris/July_26|July 26]]):<br> | ||
| + | - 50% wt rfp / 50% dapA- (colomn 1)<br> | ||
| + | - 90% wt rfp / 10% dapA- (colomn 2)<br> | ||
| + | - 95% wt rfp / 5% dapA- (colomn 3)<br> | ||
| + | - 97% wt rfp / 3% dapA- (colomn 4)<br> | ||
| + | |||
| + | We took a sample (100µL) each hour, centrifuge it at 10000rpm 5 minutes to concentrate the cells and spread on slide (see [[Paris/PROTOCOLS#Fluorescent_single_cells_visualisation|Protocols]]). Then we looked under microscope. | ||
| + | |||
| + | [[Image: rfp_dap-_08032007.jpg|center|600px]] | ||
| + | |||
| + | The dapA- bacteria look surviving. They act as parasite of wt rfp cells because wt cells do not need dapA- cells. So this is just a preliminary experiment because in our system, cells able to multiply need cells unable because of high dapA secretion. This was to see how many wt cells are needed to dapA cells to survive. | ||
| + | |||
| + | == TOPO cloning == | ||
| + | |||
| + | Cloning using TOPO protocol of the following purification of PCR products<br> | ||
| + | |||
| + | {| | ||
| + | |- style="background: #ccccff; text-align: center;" | ||
| + | |width=5%| Number | ||
| + | |width=10%| Name | ||
| + | |width=15%| Oligo F | ||
| + | |width=15%| Oligo R | ||
| + | |width=10%| Matrix | ||
| + | |width=5%| Length | ||
| + | |width=25%| Comments | ||
| + | |- style="background: #cccccc;" | ||
| + | | style="background: #ccffcc;" |P6 | ||
| + | |Lox66-DapASubtilis | ||
| + | |O8 Lox66-DapASubt-F | ||
| + | |O9 DapASubt-R | ||
| + | |B.Subtilis 168 | ||
| + | |979 | ||
| + | | | ||
| + | |- style="background: #cccccc;" | ||
| + | | style="background: #ccffcc;" |P7 | ||
| + | |Lox71-FtsZ | ||
| + | | O1 Lox71-FtsZ-F | ||
| + | | O2 FtsZ-R | ||
| + | |MG1655 | ||
| + | |1260 | ||
| + | |EcoRI site in FtsZ not mutated | ||
| + | |- style="background: #cccccc;" | ||
| + | | style="background: #ccffcc;" |P8 | ||
| + | |RBS-DapAColi | ||
| + | | O20 RBS-DapAColi-F | ||
| + | | O7 DapAColi-R | ||
| + | | MG1655 | ||
| + | |952 | ||
| + | |B0030-DapAColi | ||
| + | |} | ||
Latest revision as of 17:53, 7 October 2007
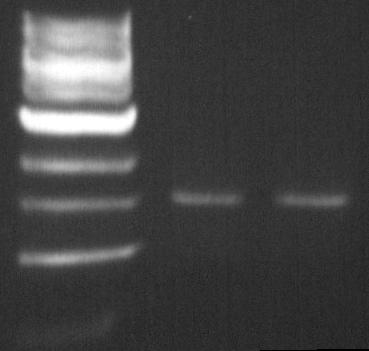
DGAT biobrick: EcoR1-Pst1 digestion
The double digestion allows us to insert the biobrick into biobrick plasmid.
The Pst1 digestion allows us to see if the mutagenesis worked (dgat gene not digested).
- Material:
- DNA (PCR product DGAT deltaPst-1 biobrick from August 2): 200ng = 20µL
- BSA: 0.5µL
- NEBuffer EcoR1: 5µL
- EcoR1 enzyme: 1µL
- Pst-1 enzyme: 1µL
- ddH2O
- Protocol:
- 2 hours at 37°C
- Purification
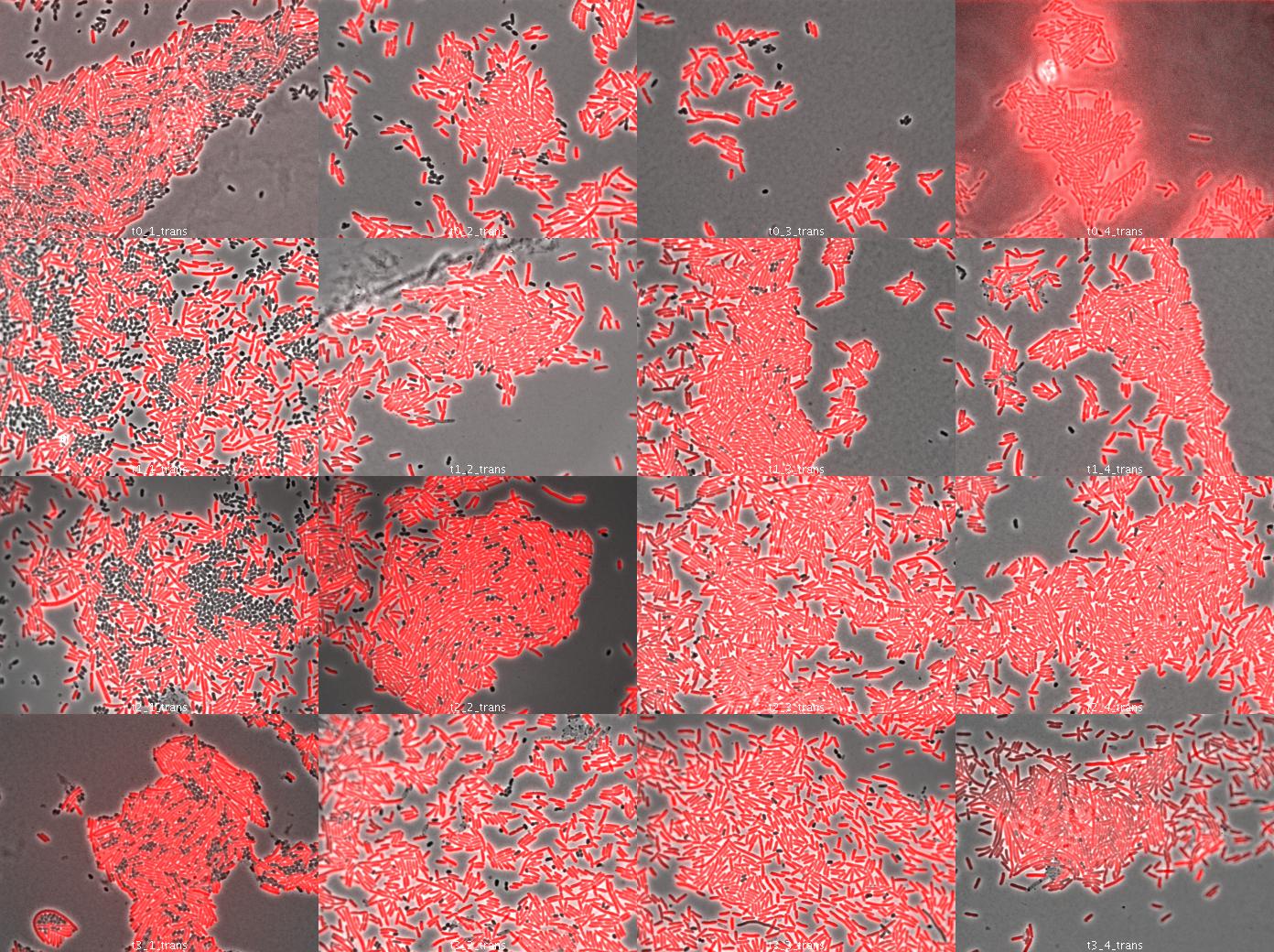
Dap complementation experiment
We used wild type E.coli transformed by part J23107 (RFP) and dapA- E.coli (w121) auxotroph to DAP. E.coli transformed by part J23107 (RFP) is able to synthesize DAP and export it. We suppose dap- E.coli could be feed by wt E.coli.
We tried to observe kinetics by fluorescence microscopy. We let separately in culture overnight dapA- E.coli (LB/erythromycine/DAP) and rfp wt E.coli (LB ampicillin). The day after, we diluted both liquid culture 1/200 to have exponential culture. At OD = 0.7 (dapA-) and OD = 0.5 (wt rfp), we mix both population in different condition culture (We used preliminary results (July 26):
- 50% wt rfp / 50% dapA- (colomn 1)
- 90% wt rfp / 10% dapA- (colomn 2)
- 95% wt rfp / 5% dapA- (colomn 3)
- 97% wt rfp / 3% dapA- (colomn 4)
We took a sample (100µL) each hour, centrifuge it at 10000rpm 5 minutes to concentrate the cells and spread on slide (see Protocols). Then we looked under microscope.
The dapA- bacteria look surviving. They act as parasite of wt rfp cells because wt cells do not need dapA- cells. So this is just a preliminary experiment because in our system, cells able to multiply need cells unable because of high dapA secretion. This was to see how many wt cells are needed to dapA cells to survive.
TOPO cloning
Cloning using TOPO protocol of the following purification of PCR products
| Number | Name | Oligo F | Oligo R | Matrix | Length | Comments |
| P6 | Lox66-DapASubtilis | O8 Lox66-DapASubt-F | O9 DapASubt-R | B.Subtilis 168 | 979 | |
| P7 | Lox71-FtsZ | O1 Lox71-FtsZ-F | O2 FtsZ-R | MG1655 | 1260 | EcoRI site in FtsZ not mutated |
| P8 | RBS-DapAColi | O20 RBS-DapAColi-F | O7 DapAColi-R | MG1655 | 952 | B0030-DapAColi |