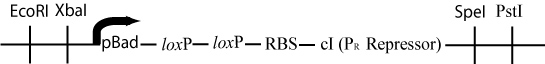Davidson Missouri W
From 2007.igem.org
Wideloache (Talk | contribs) (→'''Proposed Construct with Cre/loxP''') |
Wideloache (Talk | contribs) (→'''Proposed Construct with Cre/loxP''') |
||
| Line 85: | Line 85: | ||
In order to determine if a Hamiltonian Path exists in the directed graph above, we propose a plasmid design similar to that shown below: | In order to determine if a Hamiltonian Path exists in the directed graph above, we propose a plasmid design similar to that shown below: | ||
| + | |||
[[Image:Plasmid before Excision.jpg]] | [[Image:Plasmid before Excision.jpg]] | ||
| - | This cartoon represents a solved arangement of our proposed construct. Between the two pairs of BioBrick restriction enzyme sites lies a region containing eight flippable DNA fragments, each flanked by hixC sites (represented by the black, jagged rectangles). Each node of the graph represents one of the following genes: GFP (Green Flourescent Protein), Kanamycin Resistance, cre, pir200, | + | This cartoon represents a solved arangement of our proposed construct. Between the two pairs of BioBrick restriction enzyme sites lies a region containing eight flippable DNA fragments, each flanked by hixC sites (represented by the black, jagged rectangles). Each node of the graph represents one of the following genes: GFP (Green Flourescent Protein), Kanamycin Resistance, cre, pir200, and a transcription terminator. Each edge of the graph is included in the construct between two hixC sites. In the presence of Hin protein, flipping of the edges will produce random walks through the graph. When flipped into the correct orientation and located upstream of any transcription terminators, a given gene will be transcribed (denoted by a bold outline). In the solved arrangement shown above, a Hamiltonian Path is determined to exist in the graph because all genes are expressed, and no extra edges exist in the coding region. Extra edges in the coding region can be detected by gel electrophoresis. |
The four reporter genes shown above will produce three distinct phenotypes. GFP will cause the E. Coli cells to glow green. Kanamycin resistance will allow the cells to grow in the presence of the antibiotic, Kanamycin. When the cre gene is in the solved orientation, it produces the Cre protein. It has been demonstrated [inster citation] that Cre can bind to two separated loxP sites and excise the DNA in the middle, forming a new plasmid. If Cre was produced, then the plasmid would be cut at its loxP sites, and produce the two plasmids shown below. | The four reporter genes shown above will produce three distinct phenotypes. GFP will cause the E. Coli cells to glow green. Kanamycin resistance will allow the cells to grow in the presence of the antibiotic, Kanamycin. When the cre gene is in the solved orientation, it produces the Cre protein. It has been demonstrated [inster citation] that Cre can bind to two separated loxP sites and excise the DNA in the middle, forming a new plasmid. If Cre was produced, then the plasmid would be cut at its loxP sites, and produce the two plasmids shown below. | ||
| Line 98: | Line 99: | ||
[[Image:Orig Plasmid after excision.jpg]] | [[Image:Orig Plasmid after excision.jpg]] | ||
| - | The original plasmid has now lost all of the DNA between the two loxP sites. The pBad promoter | + | The original plasmid has now lost all of the DNA between the two loxP sites. The pBad promoter (initially upstream of the first loxP site) has been moved just upstream of ribsosomal binding site and the cI gene (initially downstream of the second loxP site). This rearrangement will cause the transcription of cI, a gene that represses the P(r) promoter in the lamda phage. |
== '''Resources / Citations'''== | == '''Resources / Citations'''== | ||
Revision as of 22:54, 8 June 2007
===Davidson & Missouri Western Team Logos, iGEM2006===
Contents |
Team Meeting Notes
Western Meeting Notes 051407 to Present [1]
Students
• Will DeLoache, Junior Biology Major, [2]
• Oyinade Adefuye, Senior Biology Major, [3]
• Jim Dickson, Junior Math and Economics Major, [4]
• Amber Shoecraft, Math Major, [5]
• Andrew Martens, Senior Biology Major, [6]
• Michael Waters, Sophomore Biology Major, [7]
• Jordan Baumgardner, Junior Biology, Biochemistry/Molecular Biology Major, [8]
• Ryan Chilcoat, Junior Biology Major (Health Sciences), [9]
• Tom Crowley, Senior Biochemisty/Molecular Biology Major, [10]
• Lane H. Heard, Central High School graduate, [11]
• Nickolaus Morton, Junior Chemistry Major, [12]
• Michelle Ritter, Junior Mathematics Major, [13]
• Jessica Treece, Junior Biology Major (Health Sciences), [14]
• Matthew Unzicker, Senior Biochemistry/Molecular Biology Major, [15]
• Amanda Valencia, Senior Biochem/Molecular Biology Major, [16]
Faculty
• Malcolm Campbell [http://www.bio.davidson.edu/people/macampbell/macampbell.html], Professor, Department of Biology, [17]
• Karmella Haynes [http://www.bio.davidson.edu/people/kahaynes/kahaynes.html], Visiting Assistant Professor, Department of Biology, [18]
• Laurie Heyer [http://www.davidson.edu/math/heyer/], Associate Professor, Department of Mathematics, [19]
Shipping Address: Malcolm Campbell, Biology Dept. Davidson College, 209 Ridge Road, Davidson, NC 28036 [(704) 894-2692]
• Todd Eckdahl [http://staff.missouriwestern.edu/~eckdahl/], Professor, Department of Biology, [20]
• Jeff Poet [http://staff.missouriwestern.edu/~poet/], Assistant Professor, Department of Computer Science, Mathematics, and Physics, [21]
Shipping Address: Todd Eckdahl, Biology Department, Missouri Western State University, 4525 Downs Drive, Saint Joseph, MO, 64507 [(816) 271-5873]
Project Overview
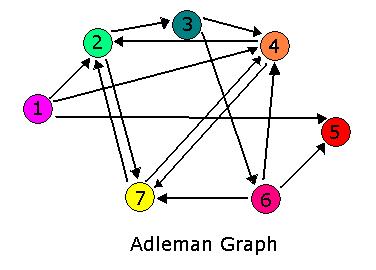
Hamiltonian Path Problem As a part of iGEM2006, a combined team from Davidson College and Missouri Western State University reconstituted a hin/hix DNA recombination mechanism which exists in nature in Salmonella as standard biobricks for use in E. coli. The purpose of the 2006 combined team was to provide a proof of concept for a bacterial computer in using this mechanism to solve a variation of The Pancake Problem from Computer Science. This task utilized both biology and mathematics students and faculty from the two institutions.
For 2007, we continue our collaboration and our efforts to manipulate E. coli into mathematics problem solvers as we refine our efforts with the hin/hix mechanism to explore another mathematics problem, the Hamiltonian Path Problem. This problem was the subject of a groundbreaking paper by Adelman in 1994 (citation below) where a unique Hamiltonian path was found in vitro for a particular directed graph on seven nodes. We propose to make progress toward solving the particular problem in vivo.
Proposed Construct with Cre/loxP
In order to determine if a Hamiltonian Path exists in the directed graph above, we propose a plasmid design similar to that shown below:
This cartoon represents a solved arangement of our proposed construct. Between the two pairs of BioBrick restriction enzyme sites lies a region containing eight flippable DNA fragments, each flanked by hixC sites (represented by the black, jagged rectangles). Each node of the graph represents one of the following genes: GFP (Green Flourescent Protein), Kanamycin Resistance, cre, pir200, and a transcription terminator. Each edge of the graph is included in the construct between two hixC sites. In the presence of Hin protein, flipping of the edges will produce random walks through the graph. When flipped into the correct orientation and located upstream of any transcription terminators, a given gene will be transcribed (denoted by a bold outline). In the solved arrangement shown above, a Hamiltonian Path is determined to exist in the graph because all genes are expressed, and no extra edges exist in the coding region. Extra edges in the coding region can be detected by gel electrophoresis.
The four reporter genes shown above will produce three distinct phenotypes. GFP will cause the E. Coli cells to glow green. Kanamycin resistance will allow the cells to grow in the presence of the antibiotic, Kanamycin. When the cre gene is in the solved orientation, it produces the Cre protein. It has been demonstrated [inster citation] that Cre can bind to two separated loxP sites and excise the DNA in the middle, forming a new plasmid. If Cre was produced, then the plasmid would be cut at its loxP sites, and produce the two plasmids shown below.
The DNA fragment located between the two loxP sites has now been excised and formed into a new plasmid. This solved plasmid is resistant to kanamycin (due to the solved orientation of the kanamycin resistance gene) and contains the gamma-ori origin of replication. It has been shown [insert citation] that the pi-protein (transcribed by the pir200 gene) initiates amplification of plasmids with the gamma-ori origin of replication. Therefore, only when the pir200 gene is in the solved orientation will the excised plasmid will be amplified.
The original plasmid has now lost all of the DNA between the two loxP sites. The pBad promoter (initially upstream of the first loxP site) has been moved just upstream of ribsosomal binding site and the cI gene (initially downstream of the second loxP site). This rearrangement will cause the transcription of cI, a gene that represses the P(r) promoter in the lamda phage.
Resources / Citations
Cool site for Breakfast [http://www.cut-the-knot.org/SimpleGames/Flipper.shtml]
Karen Acker's paper describing GFP and TetA(c) with Hix insertions [http://www.bio.davidson.edu/Courses/Immunology/Students/spring2006/Acker/Acker_finalpaperGFP.doc]
Bruce Henschen's paper describing one-time flippable Hix sites [http://www.bio.davidson.edu/Courses/genomics/2006/henschen/Bruce_Finalpaper.doc]
Intro to Hamiltonian Path Problem and DNA [http://www.ams.org/featurecolumn/archive/dna-abc2.html]
Adelman, LM. Molecular Computation of Solutions To Combinatorial Problems. Science. 11 November 1994. Vol. 266. no. 5187, pp. 1021 - 1024
Literature and Registry Research
Registry Search for Possible Promoters:
BBa_J24669 --- arabinose induced
BBa_R0082 --- Is upstream of the ompC porin gene
BBa_R0074 --- Penl regulated
BBa_I14017 --- P(Rhl)
BBa_I14018 --- P (Bla) --> amp resistance
BBa_J3902 --- Pr Fe (Pl + Pll rus operon)
BBa_R0077 --- CinR --> thought to have own terminator
BBa_R0078 --- CinR (no RBS)
BBa_R0062 --- luxR & HSL regulated -- luxpR
Possibly the use of Constitutive Promoter Family Members to strengthen other promoters.
Literature Search for Polycistronic Genes on Plasmids
Sol Operon http://www.jstage.jst.go.jp/article/bbb/71/1/58/_pdf
Transfer (tra) Operon http://www.pubmedcentral.nig.gov/picrender.fcgi?artid=1347297&blobtype=pdf
Oligopeptide Permease (opp) http://www.pubmedcentral.nig.gov/picrender.fcgi?artid=1087318&blobtype=pdf