ETHZ/Biology
From 2007.igem.org
Stefan Luzi (Talk | contribs) (→Reporting Subsystem) |
Stefan Luzi (Talk | contribs) (→Reporting Subsystem) |
||
| Line 27: | Line 27: | ||
==== Reporting Subsystem ==== | ==== Reporting Subsystem ==== | ||
| - | <p>There are four reporters in the system. CFP (more precisely: enhanced CFP, that is ECFP) and YFP (more precisely: enhanced YFP, that is EYFP) are active during the learning phase of the system and show which chemical the system is exposed to during | + | <p>There are four reporters in the system. CFP (more precisely: enhanced CFP, that is ECFP) and YFP (more precisely: enhanced YFP, that is EYFP) are active during the learning phase of the system and show which chemical the system is exposed to during learning, whereas all four reporters (the latter and GFP and RFP) are active during the recognition phase and show if the system is exposed to the same chemical as in learning or not. |
More specifically, the YFP production is regulated with help of two operator sites controlled by cI and aTc (TetR inhibitor). cI inhibits YFP production and aTc induces it. Therefore, YFP is synthesized when the system is exposed to only aTc but not to cI. The production of the other fluorescent proteins is regulated in a similar manner:<br> CFP gets produced when the system is exposed to only IPTG but not to p22cII<br> | More specifically, the YFP production is regulated with help of two operator sites controlled by cI and aTc (TetR inhibitor). cI inhibits YFP production and aTc induces it. Therefore, YFP is synthesized when the system is exposed to only aTc but not to cI. The production of the other fluorescent proteins is regulated in a similar manner:<br> CFP gets produced when the system is exposed to only IPTG but not to p22cII<br> | ||
GFP gets produced when the system is exposed to only IPTG but not to cI<br> | GFP gets produced when the system is exposed to only IPTG but not to cI<br> | ||
Revision as of 18:12, 23 October 2007

On this page, you can find an analysis of the function of our system, its biological design, and a list of the parts that make up the system. Are you interested in constructing educatETH E.coli in your lab? Then under Lab Notes, you can find the ingredients and equipment we used, the electronic version of our lab notebook and a presentation of all the difficulties that we encountered. If you are also interested in how educatETH E.coli was modeled and simulated outside the lab, please visit the modeling and simulations web pages.
Introduction
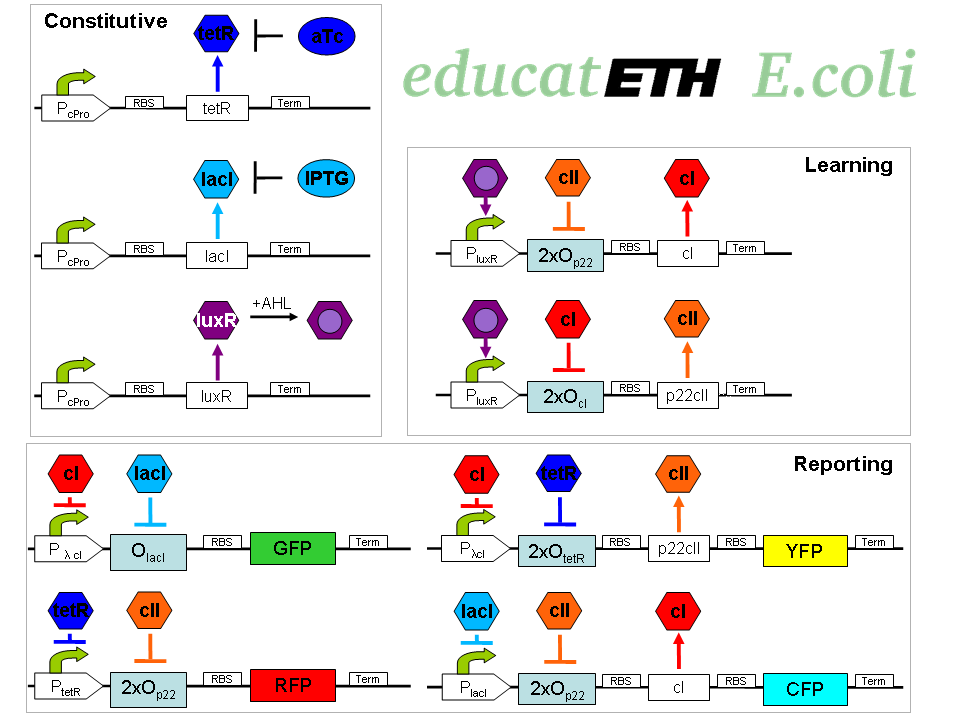
educatETH E.coli is a system which can distinguish between [http://openwetware.org/wiki/ATc anhydrotetracycline (aTc)] and [http://openwetware.org/wiki/IPTG Isopropyl-beta-D-thiogalactopyranoside (IPTG)] based on a previous learning phase conducted with the same chemicals and the help of [http://partsregistry.org/Acyl-HSLs AHL]. It is composed of three subsystems: the subsystem of constitutively produced proteins, the learning subsystem and the reporting subsystem. The constitutively produced proteins (LacI, TetR and LuxR) control the learning subsystem. At the core of the latter there exists an extended version of the original toggle switch found in [1]. That is, a multi-inducible toggle switch. The main difference is reflected in the use of double promoters, so that the toggle switch only changes its state when both, one of the two chemicals (aTc/IPTG), and AHL are present. As AHL is only present during the learning phase, the toggle maintains its state during testing/recognition, and thus can “memorize”. AHL can therefore be seen as a training- or learning substance. In the reporting subsystem, four reporters ([http://partsregistry.org/Featured_Parts:Fluorescent_proteins fluorescent proteins]) allow supervision of (1.) the chemical the system was trained with and (2.) if the system recognizes the chemical it is being exposed to in the recognition phase as one it has been previously trained with or not.
The Complete System
The biological design of educatETH E.coli is presented in Fig. 1 and below, we clarify the function of all depicted components. (Are you interested in how the complex system of Fig. 1 was modeled? Then visit the System Modeling!)Constitutive Subsystem
The constitutively produced proteins of the system are LacI, TetR and LuxR. The LuxR part has a special function: when AHL is present, it forms a LuxR-AHL complex which acts on the learning subsystem (more on this later). For now, we will consider that AHL is absent and therefore LuxR cannot act on any subsystems. The TetR and LacI parts behave similarly: more specifically, the TetR protein in the absence of aTc inhibits the production of p22cII and LacI in the absence of IPTG inhibits the production of cI. When aTc is present, however, the p22cII production is no longer inhibited (and thus p22cII is produced). Correspondingly, cI is produced when IPTG is present.
Learning Subsystem
The learning subsystem is a toggle switch with two operator sites. The upper part of the toggle (cI production) has operator sites for the LuxR-AHL complex and p22cII (which has in turn been induced by aTc). The LuxR-AHL complex induces cI production, whereas p22cII inhibits it. The lower part of the toggle (p22cII production) has operator sites for the LuxR-AHL complex and cI (which has been induced by IPTG). In analogy to the upper part, the LuxR-AHL complex induces production of p22cII and cI inhibits it. Therefore, the switch always requires the presence of the LuxR-AHL complex in order for it to operate. Its state depends on the presence of p22cII and cI in the system, which in turn was caused through the exposure of the system to aTc and IPTG.
Reporting Subsystem
There are four reporters in the system. CFP (more precisely: enhanced CFP, that is ECFP) and YFP (more precisely: enhanced YFP, that is EYFP) are active during the learning phase of the system and show which chemical the system is exposed to during learning, whereas all four reporters (the latter and GFP and RFP) are active during the recognition phase and show if the system is exposed to the same chemical as in learning or not.
More specifically, the YFP production is regulated with help of two operator sites controlled by cI and aTc (TetR inhibitor). cI inhibits YFP production and aTc induces it. Therefore, YFP is synthesized when the system is exposed to only aTc but not to cI. The production of the other fluorescent proteins is regulated in a similar manner:
CFP gets produced when the system is exposed to only IPTG but not to p22cII
GFP gets produced when the system is exposed to only IPTG but not to cI
RFP gets produced when the system is exposed to only aTc but not to p22cII
System Phases
The system operation is divided into two main phases: the training phase and the testing phase. The training phase itself is also subdivided into two phases: seeing and memorizing. During seeing, the system is first exposed to one of the two chemicals it is designed to recognize (aTc and IPTG). AHL is then added and the system’s internal toggle switch reaches a steady state. During memorizing, the chemical used during seeing is removed and only AHL is retained. This maintains the toggle switch to its acquired steady state, which is reported with YFP (if aTc was seen) or CFP (if IPTG was seen). During the testing phase, the system is exposed to any of the two chemicals (aTc or IPTG), with AHL present. By comparing its toggle switch state with the effect of the newly introduced chemical, the system shows a different response if it has previously been exposed to this chemical and reports with the same XFP as in the training phase (YFP for aTc, CFP for IPTG) or if it recognizes a different chemical and reports with a different XFP (GFP for trained with aTc and recognizing IPTG, RFP for trained with IPTG and recognizing aTc).The following table presents all possible paths that may be taken by the system during all phases of operation according to the external stimuli.
| aTc | IPTG | AHL | p22cII | cI | Reporting | |
|---|---|---|---|---|---|---|
| Start | ||||||
| no input | no | no | no | no | no | no |
| Learning | ||||||
| Trained with aTc | yes | no | no | yes | no | YFP |
| Trained with IPTG | no | yes | no | no | yes | CFP |
| Memorizing | ||||||
| Trained with aTc | yes | no | yes | yes | no | YFP |
| Trained with IPTG | no | yes | yes | no | yes | CFP |
| Recognition | ||||||
| Trained with aTc Tested with aTc | yes | no | yes | yes | no | YFP |
| Trained with aTc Tested with IPTG | no | yes | yes | yes | no | GFP |
| Trained with IPTG Tested with IPTG | no | yes | yes | no | yes | CFP |
| Trained with IPTG Tested with aTc | yes | no | yes | no | yes | RFP |
System Parts
educatETH E.coli consists of 11 parts that can be synthesized independently (want to know how this is done in the lab? Then visit our In the Lab page!) Please note that four of them (4,5 and 8,9) form together two functional system units. They have been separated to ensure comparable part lengths and thus enable easier introduction into plasmids. A detailed list of the used parts can be found here (due to many pictures, it takes some time to load).
References
[http://www.nature.com/nature/journal/v403/n6767/abs/403339a0.html [1] Gardner TS, Cantor CR and Collins JJ] "Construction of a genetic toggle switch in Escherichia coli", Nature 403:339–342, 2000
To Do
New
<p>- Update and correct parts in parts list. Write better in a table
- Update and correct full system scheme
- What is the proof of concept mentioned?
- Are you sure about the reporters? This is not what I understood. I though YFP and CFP were also during testing. Yes! I'm still updating the text :-)
- I don't think it is wise to make p22cII in P22 cII, since we have in the first way all over the place. Stefan: The problem is that the correct notation is P22 cII and I already used it in all the part describtions. But I could also change it back, no worries!
- Try to improve table with system phases. It doesn't look so nice...
- In the memorizing phase, is there color or not? - Nope, there is no color - There are colors, however they will disappear within 2-3 hours.
- We need to put the complete spaghetti system as well.
- Check my terminology (operator sites etc)
- Put Stefan's updated part on epigenetics
- Fill in table completely, make it more reading-friendly