Paris/Cell auto
From 2007.igem.org
(→'''''Parameters''''') |
(→Spatial simulation) |
||
| Line 3: | Line 3: | ||
<br> | <br> | ||
==Spatial simulation== | ==Spatial simulation== | ||
| - | We try with this work to characterize the diffusion of the DAP and the effect on the cells. We have a | + | We try with this work to characterize the diffusion of the DAP and the effect on the cells. We have a lawn of bacteries with germinal cells and some somatic cells.<br> |
| + | |||
=='''''We make some hypothesis:'''''== | =='''''We make some hypothesis:'''''== | ||
Revision as of 21:31, 23 October 2007
Contents |
Spatial simulation
We try with this work to characterize the diffusion of the DAP and the effect on the cells. We have a lawn of bacteries with germinal cells and some somatic cells.
We make some hypothesis:
We work with a constant population (no death and no division), we can imagine that we are a stationary phase or between to division cycle.
The differentiation is DAP dependent, it's append when the cell as enough DAP to evolve but not enough to divide.
The DAP is made in bacteria S, the production rate is the difference between the total production and self consummation
The DAP can be under two types intra/extra cellular (DAPi/DAPe)
The DAP is consumed in bacteria G
We have 3 entities in our model
Bact it has a concentration internal of DAP (DAPi) and external (DAPe). It's a cell in our automaton
BactS is a Bact which produce DAPi
BactG is a Bact which consume DAPi
We produce this set of rules
For bactS
*DAPe = DAPe + DAPe_diffused_in_neighborhood + DAPi_diffused_from_the_BactS
*DAPi= DAPi +DAPi_produced - DAPi_diffused_from_the_BactS
For BactG
*DAPe=DAPe + DAPe_diffused_in_neighborhood - DAPi_diffused_from_the_BactG
*DAPi= DAPi -DAPi_consummate + DAPi_diffused_from_the_BactG
*BactG = if minimal DAPi for differentiation < DAPi < maximal DAPi for differentiation BactS else stay BactG
Initial state
We use a 30x30 cells automaton.
All cells are BactG excepted 4 BactS which are placed randomly on the automaton
Parameters
We have 8 parameters and we can add noise for each of them.
In BactS:
- Dap export
- Dap import
- Dap production
In BactG:
- Dap export
- Dap import
- Dap consummation
- Minimal Dap needed for differentiation
- Maximal Dap needed for differentiation
Output
The output is two animated pictures one show the differentiation the other the diffusion of DAPe

- The first picture show the diffusion of DAP
- We can see a front wave in light blue after that there is a dark blue area in which the systeme is stable the concentration doesn't evolve.
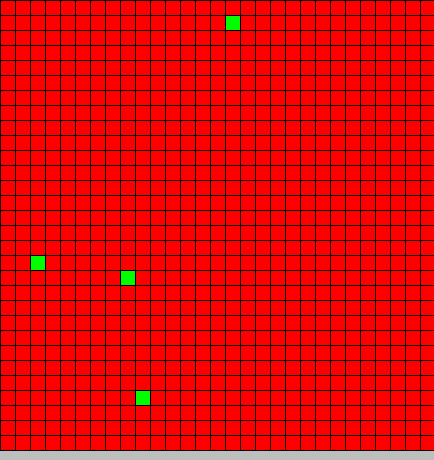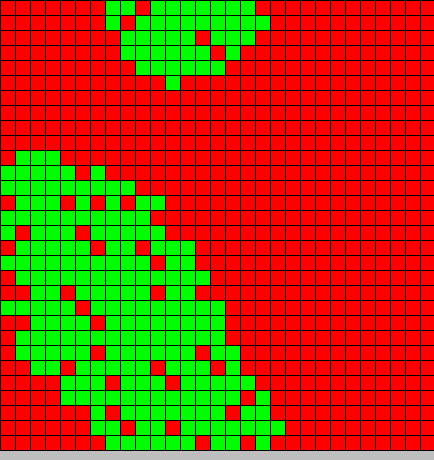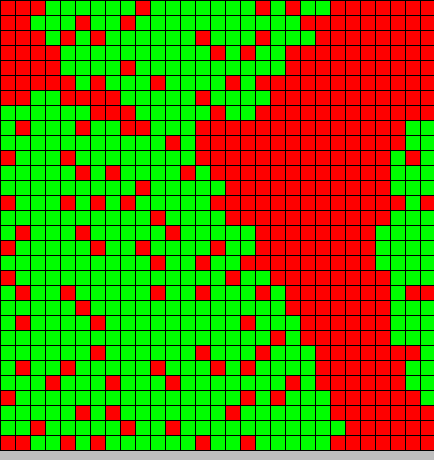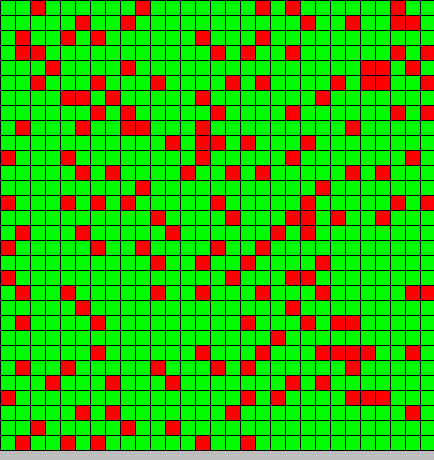
- The second picture show the differentiation
- Red BactG
- Green BactS
- The differentiation follow the wave front
After playing with the parameters, we can deduct 2 important things:
- The inhibition most be strong and effective (we play with the minimal and maximal value of DAP for differentiation)
- if it isn't the case the system collapse all the bactG stay BactG if the inhibition is too strong or switch to BactS if the inhibition is not enough strong.
- The production and diffusion of DAP will be a critical factor
- The DAP has to be produce then he will be exported, it will diffuse in the medium and will be imported
- There is no proof of a special system to import or export DAP, so for each step there is a large amount of DAP lost.
