See here
From 2007.igem.org
| (3 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| - | As indicated earlier, in order for the modified SMB to behave as a secure source of sterile somatic cells, | + | As indicated earlier, in order for the modified SMB to behave as a secure source of sterile somatic cells, three events should be initiated upon activation of a switch. This could be done in the following manner: |
| - | * EVENT 1 | + | * EVENT 1 |
| - | * EVENT 2 | + | In order to achieve massive induction of recombination,, we could add inducible promoter in regulation of Cre expression (pBad for example). It could allow us to differentiate every cell into somatic cell. The generation of a substantial number of somatic cells should be possible if we let the system work alone (positive feedback by pDAPA). And once this number is enough we could add arabinose to express Cre in every germ cell (long and strong induction), leading to massive G to S differentiation. |
| + | But a certain proportion of G cells would however persist. | ||
| + | |||
| + | * EVENT 2 | ||
| + | |||
| + | The solution to second challenge could be coupling the high recombination switch to a retarded suicide switch in the gem line cells. This could be the expression of ccdB suicide gene in the germ line cells. | ||
This could be the opportunity to practically apply one of the founder principles of synthetic biology: modularity in systems assembly. | This could be the opportunity to practically apply one of the founder principles of synthetic biology: modularity in systems assembly. | ||
| Line 14: | Line 19: | ||
Using such a time delay circuit could allow using a single signal to induce event 1, followed after a delay by event 2. | Using such a time delay circuit could allow using a single signal to induce event 1, followed after a delay by event 2. | ||
| + | * EVENT 3 | ||
| + | We could also be able to screen the somatic cells by the size. Indeed S cells that have lost FtsK expression present a strange phenotype: they grows without dividing. We also could keep big cells (Cell sorting). The threshold should be determined by studying the size repartition of germ cells and somatic cells to improve sensitivity and specificity of our test. | ||
| + | |||
| + | [[Image: security scheme.jpg|400px]] | ||
[[Image: security device.jpg|right|500px]] | [[Image: security device.jpg|right|500px]] | ||
| - | + | ||
Latest revision as of 22:54, 24 October 2007
Back to Paris/Applications
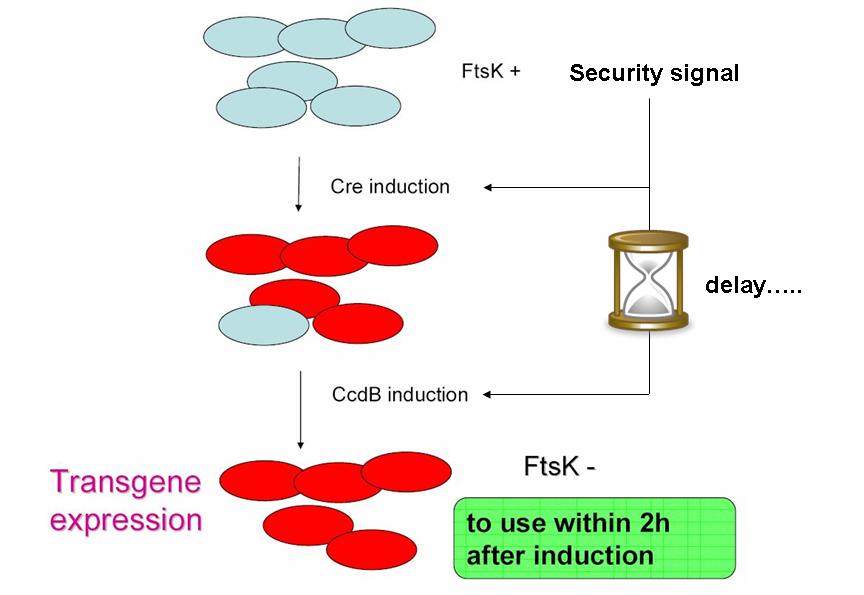
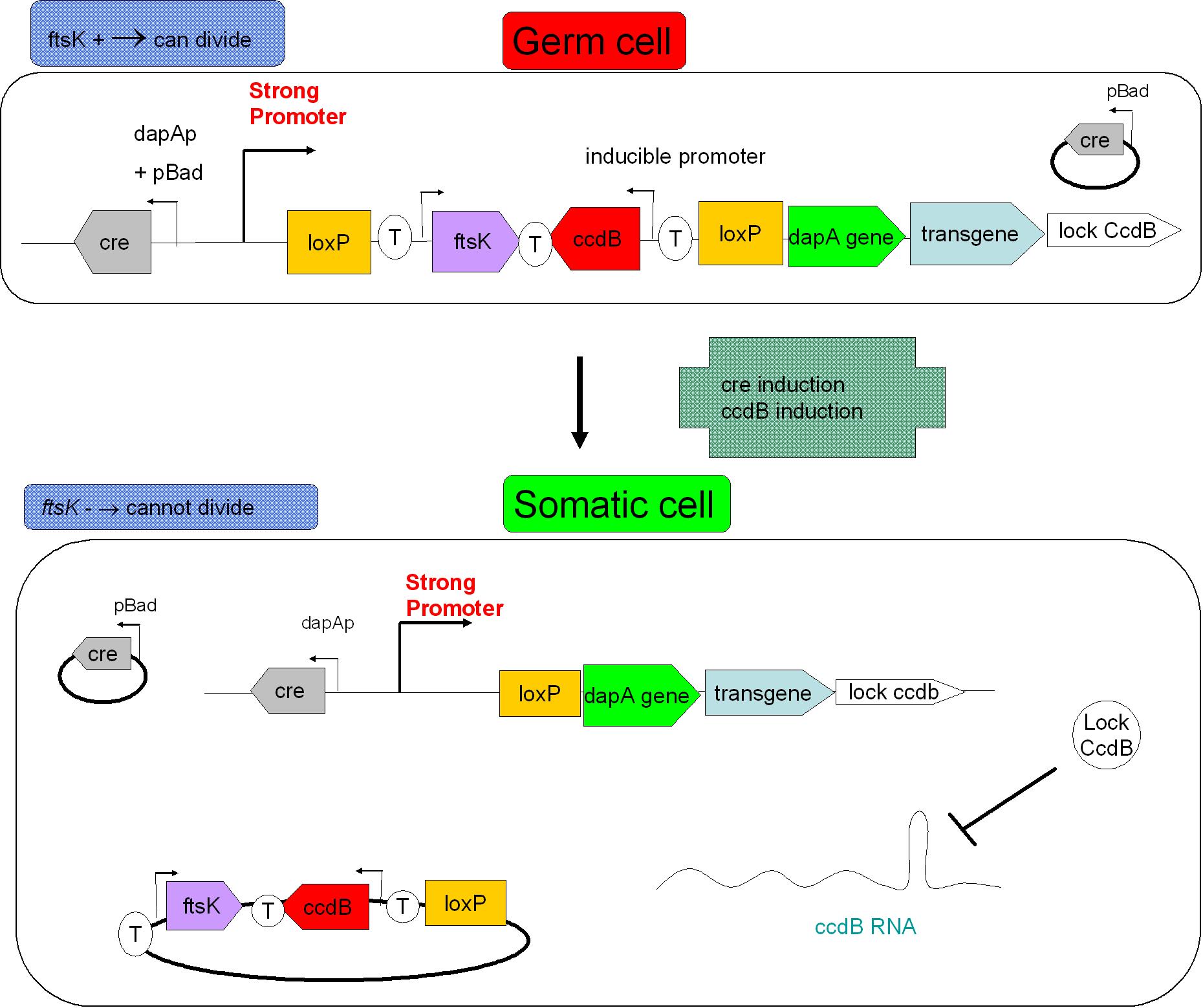
As indicated earlier, in order for the modified SMB to behave as a secure source of sterile somatic cells, three events should be initiated upon activation of a switch. This could be done in the following manner:
- EVENT 1
In order to achieve massive induction of recombination,, we could add inducible promoter in regulation of Cre expression (pBad for example). It could allow us to differentiate every cell into somatic cell. The generation of a substantial number of somatic cells should be possible if we let the system work alone (positive feedback by pDAPA). And once this number is enough we could add arabinose to express Cre in every germ cell (long and strong induction), leading to massive G to S differentiation. But a certain proportion of G cells would however persist.
- EVENT 2
The solution to second challenge could be coupling the high recombination switch to a retarded suicide switch in the gem line cells. This could be the expression of ccdB suicide gene in the germ line cells.
This could be the opportunity to practically apply one of the founder principles of synthetic biology: modularity in systems assembly.
Indeed, a time delay circuit has already been described, constructed & theoretically studied (Sara Hooshangi et al. 2005, Sara Hooshangi et al. 2006 and Juan M. Pedraza et al. 2006). Using such a time delay circuit could allow using a single signal to induce event 1, followed after a delay by event 2.
- EVENT 3
We could also be able to screen the somatic cells by the size. Indeed S cells that have lost FtsK expression present a strange phenotype: they grows without dividing. We also could keep big cells (Cell sorting). The threshold should be determined by studying the size repartition of germ cells and somatic cells to improve sensitivity and specificity of our test.
Ref.
- Ultrasensitivity and noise propagation in a synthetic transcriptional cascade
Sara Hooshangi, Stephan Thiberge, and Ron Weiss PNAS � March 8, 2005 � vol. 102 � no. 10 � 3581–3586
- Noise Propagation in Gene Networks
Juan M. Pedraza and Alexander van Oudenaarden SCIENCE VOL 307 25 MARCH 2005
- The effect of negative feedback on noise propagation in transcriptional gene networks
Sara Hooshangi and Ron Weiss CHAOS 16, 026108 (2006)