Tristable
From 2007.igem.org
(Difference between revisions)
Norrishung (Talk | contribs) (→Email Correspondances about Tri-Stable Switch) |
|||
| (36 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
<!-- | <!-- | ||
| Line 15: | Line 16: | ||
<html> | <html> | ||
<style> | <style> | ||
| + | a:link{ | ||
| + | |||
| + | text-decoration:none; | ||
| + | |||
| + | color:#4b8ada | ||
| + | |||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | a:visited{ | ||
| + | |||
| + | text-decoration:none; | ||
| + | |||
| + | color:#4b8ada | ||
| + | |||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | a:active{ | ||
| + | |||
| + | text-decoration:none; | ||
| + | |||
| + | color:#4b8ada | ||
| + | |||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | a:hover{ | ||
| + | |||
| + | text-decoration:underline; | ||
| + | |||
| + | color:#4b8ada | ||
| + | |||
| + | } | ||
body { | body { | ||
margin:0px 0px 0px 20px | margin:0px 0px 0px 20px | ||
| Line 68: | Line 106: | ||
| - | + | '''Click on the title to learn more about each section.''' | |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | + | =[[/Intro to Tristable | Tri-Stable Toggle Switch]]= | |
| + | [[Image:Tristable_Toggle_Switch_2007.jpg|thumb|right]]A trinary memory unit. A genetic circuit. A proof of concept. Here is the man-made architecture of the switch and the natural context of our parts. | ||
| - | |||
| Line 86: | Line 120: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| + | =[[/Modeling | Tri-Stable Model]]= | ||
| + | [[Image:beta values.png|thumb|right]]While Gardner et al created a mathematical model and a genetic switch in the Bistable paper, one was not used to design and improve upon the other. Here we extend our model to the Tristable system and discuss what certain parameters mean in terms of DNA and Proteins, Production, Repression, etc. | ||
| - | |||
| - | |||
| Line 103: | Line 133: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| + | =[[/Testing Constructs | Testing Constructs]]= | ||
| + | [[Image:betaTest.png|thumb|right]]The dirty side of the switch. Testing constructs that will enable us to determine our constants in absolute terms and apply a mathematical basis to changes we make on the Tri-Stable Switch architecture. | ||
| Line 118: | Line 146: | ||
| + | =[[/Appendix | Appendix]]= | ||
| + | More information about where we are going and where we have been. | ||
| Line 124: | Line 154: | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- | ||
| + | =Tri-stable Toggle Switch= | ||
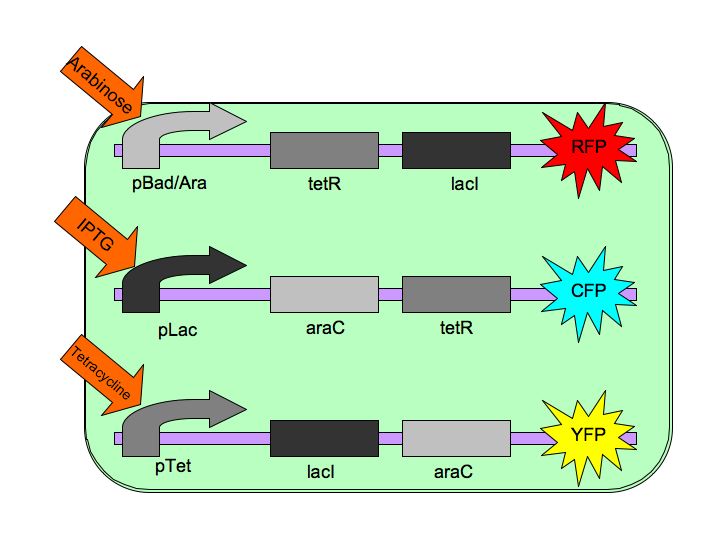
| + | The Tri-Stable switch three distinct and stable outputs in response to three distinct inputs. These three inputs are three separate chemicals which will each induce one state of the switch. [[Image:Tristable_Toggle_Switch_2007.jpg|thumb|left|The Tri-stable Toggle Switch Architecture]] In order to achieve this goal, we are constructing three constructs, each of which consists of a repressible, constitutively-on promoter attached to two repressors. Specifically, our three constructs are: | ||
| + | pBAD->LacI->TetR, | ||
| + | |||
| + | pLacI->AraC->TetR and | ||
| + | |||
| + | pTet->AraC->LacI, | ||
| + | |||
| + | where [http://en.wikipedia.org/wiki/L-arabinose_operon AraC] represses pAraC/BAD, [http://en.wikipedia.org/wiki/Lac_repressor LacI] represses pLac and [http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation TetR] represses pTet. | ||
| + | |||
| + | |||
| + | |||
| + | Each of the three repressors are inactivated by one of three chemicals, the three inducer chemicals mentioned earlier. These three([http://en.wikipedia.org/wiki/Arabinose arabinose], [http://en.wikipedia.org/wiki/IPTG IPTG] (Isopropyl β-D-1-thiogalactopyranoside) and [http://en.wikipedia.org/wiki/Tetracycline Tetracycline], respectively), cause conformational changes in their respective repressor proteins which keeps them from binding to DNA in an inhibitory manner which leads to gene expression. For example, in the presence of arabinose, AraC cannot repress pAraC/BAD so LacI and TetR are produced which in turn repress pTet and pLac and the pAraC/BAD construct is turned on. | ||
| + | |||
| + | |||
| + | |||
| + | ==AraC/BAD== | ||
| + | The gene AraC, one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] | ||
| + | [[Image:Two_Dimers_of_AraC.jpg|thumb|left|Dimer structure with arabinose on the left (yellow)]] | ||
| + | [[Image:AraC_Promoters.gif|left|thumb|The left image shows the araC dimer repressing transcription, while the right conformation enables transcription]]The protein forms a dimer with and without arabinose but the structural change activates or represses the pAraC/BAD. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==LacI== | ||
| + | In nature, LacI represses pLac which promotes the LacYZA genes that metabolize lactose. Thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image:LacI_repressor.gif|thumb|left|Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. However, an inducer, such as IPTG, causes a conformation change that removes LacI from the operator site.]] Lactose causes a conformational change which inhibits LacI from binding to the operator site of pLac. Four LacI proteins form a tetramer to inhibit pLac and four inducer molecules are required to cause the full conformational change in the repressor.[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==TetR== | ||
| + | TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter of which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation] | ||
| + | The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum] | ||
| + | [[Image:Tc_bound_to_TetR.jpg|thumb|left|A tetracycline molecule binds to each of the two TetR monomers to form a dimer]] | ||
=Modeling= | =Modeling= | ||
| Line 203: | Line 282: | ||
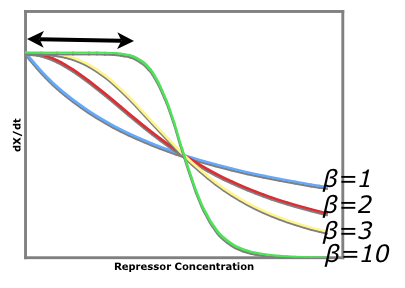
==Cooperativity of Repression== | ==Cooperativity of Repression== | ||
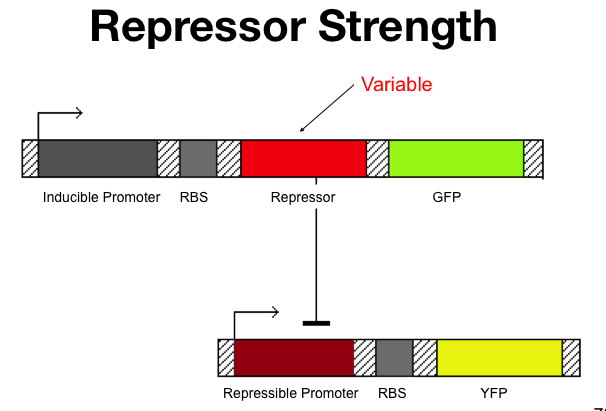
The cooperativity describes an inherent characteristic of a repressor's repression. In our system we want to know how [[Image:betaTest.png|thumb|left|The Beta test Architecture]]much increased repressor concentration will increase repression. In our model, cooperativity is an exponent, beta, so that the repressor concentration is raised to the beta ([repressor]^beta = total repression). For beta = 1, repression increases linearly with repressor concentration. With twice as much repressor there is twice the repression. For beta = 2, repression increases with the square of the concetration. Twce as much repressor leads to four times the repression. Our model predicts that our system will be more robust with greater alpha values and the tri stable region (in the graph) will be larger. Beta must be larger than one for the system to be stable. | The cooperativity describes an inherent characteristic of a repressor's repression. In our system we want to know how [[Image:betaTest.png|thumb|left|The Beta test Architecture]]much increased repressor concentration will increase repression. In our model, cooperativity is an exponent, beta, so that the repressor concentration is raised to the beta ([repressor]^beta = total repression). For beta = 1, repression increases linearly with repressor concentration. With twice as much repressor there is twice the repression. For beta = 2, repression increases with the square of the concetration. Twce as much repressor leads to four times the repression. Our model predicts that our system will be more robust with greater alpha values and the tri stable region (in the graph) will be larger. Beta must be larger than one for the system to be stable. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 214: | Line 289: | ||
==Ligand Concentration== | ==Ligand Concentration== | ||
Naturally, we don't want to add more ligand than we need. In what ever application the project might find, if we wanted to change the state again by adding a different [[Image:ligandTest.png|thumb|left|The Ligand test Architecture]]ligand, we wouldn't want excessive amounts of the first ligand floating around. | Naturally, we don't want to add more ligand than we need. In what ever application the project might find, if we wanted to change the state again by adding a different [[Image:ligandTest.png|thumb|left|The Ligand test Architecture]]ligand, we wouldn't want excessive amounts of the first ligand floating around. | ||
| + | |||
| Line 262: | Line 338: | ||
[[Media:emailwithPatrickKing.txt]] | [[Media:emailwithPatrickKing.txt]] | ||
| + | --> | ||
<html> | <html> | ||
</td> | </td> | ||
Latest revision as of 22:38, 25 October 2007

|
|||||||||||||||||