Edinburgh/DivisionPopper/References
From 2007.igem.org
JoeRokicki (Talk | contribs) |
JoeRokicki (Talk | contribs) (→References) |
||
| Line 75: | Line 75: | ||
==References== | ==References== | ||
| + | |||
| + | Hendricks EC, Szerlong H, Hill T, Kuempel P. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 2000 May;36(4):973-81. | ||
| + | |||
| + | Ptacin JL, Nollmann M, Bustamante C, Cozzarelli NR. Identification of the FtsK sequence-recognition domain. Nat Struct Mol Biol. 2006 Nov;13(11):1023-5. Epub 2006 Oct 15. | ||
| + | |||
| + | Recchia GD, Aroyo M, Wolf D, Blakely G, Sherratt DJ. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 1999 Oct 15;18(20):5724-34. | ||
| + | |||
| + | Steiner WW, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998 Dec;180(23):6269-75. | ||
| + | |||
| + | Strick TR, Quessada-Vial A. FtsK: a groovy helicase. Nat Struct Mol Biol. 2006 Nov;13(11):948-50. Erratum in: Nat Struct Mol Biol. 2007 Jun;14(6):568. | ||
Revision as of 23:16, 26 October 2007
MENU : Introduction | Background | Applications | Design&Implementation | Modelling | Wet Lab | Synthetic Biology Approach | Conclusions
Some of the biological mechanisms at the base of the Division PoPper design have been widely studied in the past and well characterized in literature (for example, the repression mechanism of Tet and lambda promoters). Others, however, are quite new and only partially understood. In particular, this is true for the dif recombinase Edinbusystem and its function resolving chromosome dimers. Given the incomplete understanding of this system as well as the novel context into which we are placing it, we were forced to make many assumptions during the design process. How will the dif recombination system react to inverting rather than excising DNA? How will shifting the system from the chromosome onto the plasmid effect the recombination system? How many inversion events should we expect per cell division? While the current literature sheds light on many of these questions, there is no substitute for an experimental answer. If the dif recombination system is to be utilized in synthetic biological constructs it must first be characterized as a device. Our project, and in particular the realization and analysis of the proof of concept system, characterizes this recombination system and its relation to cell division from a synthetic biology perspective and creates a basis for further investigations as well as a device for future constructs. In this section, we propose a brief literature review of the dif recombinase system and its functions in cell division in order to motivate and understand the results of the proof of concept device.
Contents |
A Review of the Dif Recombinase
Introduction
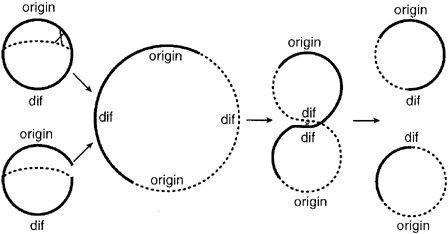
In addition to the requirement of replicating all genomic DNA once, and only once, per cell cycle and insuring that exactly one copy of each genome is distributed to the two daughter cells, E. Coli cells must also contend with the problem of chromosome dimers. Dimers form when newly replicated DNA undergoes homologous recombination with its sister chromotid. When this event occurs, the two sister chromotids link together to form one large dimer ring. In normal growth, this event occurs with a probability of at least 15% per generation. [Sister Chromatid Exchange Frequencies in Escherichia coli Analyzed by Recombination at the dif Resolvase Site- Kuempel-1998]
Figure 1: Schematic representation of the mechanisms by which chromosome dimmers arise and are resolved (figure from Kuempel 1998)
Dif Phenotype
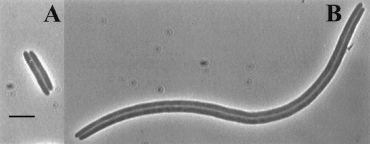
A cell without the ability to resolve dimers exhibits what is known as the dif (deletion-induced filamentation) phenotype. Even though the two sister chromosomes are linked, the cell attempts to go about replication as normal. The terminus region (the last part of the genome to undergo replication) is trapped in the vicinity of the septum (a region in the center of the cellular compartment which ultimately pinches off to form the two daughter cells). Without anyway to separate the two genomes, the cell faces one of two outcomes. Either DNA remains trapped in the septum and the cells are tethered together by their genomes and form long filaments, or the septum “guillotines” the DNA resulting in the degradation of both genomes and cell death. [Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli- Hendricks 2000]
Figure 2: Example of cells exhibiting the dif phenotype (from Hendricks 2000)
A Model For Resolution
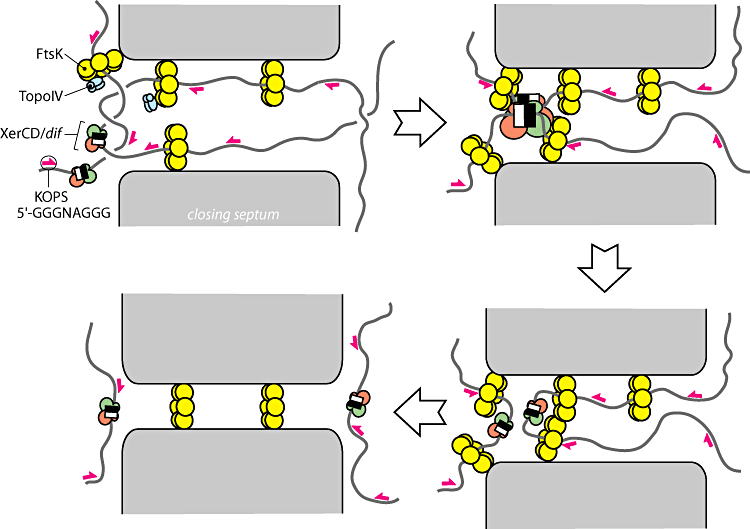
In order to resolve dimers, the cell utilizes two tyrosine recombinases XerC and XerD. These recombinases bind to a 28bp directional sequence known as a dif site. A dimer will contain two dif sites but because they will be at opposite sides of the genome even after they have been bound with recombinase they must somehow be brought into physical proximity for a recombination event to take place. To accomplish this, the cell utilizes the most powerful molecular motor yet discovered, [FtsK [FtsK: a grovy helicase Strick 2006.] Immediately before division, FtsK, assembles into many membrane bound hexamer complexes. These complexes contain a N-terminus region which anchors the complex to the septum and a C-terminus region which functions as a motor. The C-terminus motor preferentially loads onto the DNA at 8bp directional sequences known as KOPS sites. [Identification of the FtsK sequence-recognition domain-Ptacin 2006] KOPS sites are littered throughout the DNA near the terminus region like arrows pointing towards the dif sites. The motors utilize this directional information to “reel” the dif sites into the terminus. In this way, the dif sites, which can be many megabases away in terms of sequence, are brought together into close physical proximity. FtsK then works with the dif-bound recombinases to catalyze a recombination event between the two dif sites, resolving the chromosome dimer into two identical monomers.
Figure 3: Overview of the current model for dimer resolution (From Bigot-2007)
"Specifications" from Literature
[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1449051&blobtype=pdf Bloor]
- An Efficient Method of Selectable Marker Gene Excision by Xer Recombination for Gene Replacement in Bacterial Chromosomes (2006)
- This article describes a system where a gene is deleted by replacing it with an antibiotic marker. The antibiotic marker is then excised during next cell division using the naturally expressed XerCD recombinases. This is similar to what we are doing except instead of excising we are inverting.
| "The natural dif site is in the chromosome terminus region and is essential for correct chromosome segregation at cell division: deletion of dif(E. coli) results in a subpopulation of E. coli cells with a filamentous morphology (12). However, if natural dif(E. coli) is deleted, another dif(E. coli) site will not enable dimer resolution if placed outside of the 30-kb dif activity zone (DAZ) in the terminus region (6, 16). Therefore, while our method involves insertion of one or more extra dif sites at different chromosomal loci, this should not have an adverse effect on chromosome segregation, although the possibility that introduction of multiple dif sites in close proximity might result in deletion of intervening chromosomal DNA should be considered." |
- Summary: Dif sites outside of the DAZ zone shouldn't cause problems with the natural dif sites of the chromosome/plasmid
[http://www.blackwell-synergy.com/doi/full/10.1046/j.1365-2958.1998.00651.x Kuempel]
- Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli (1998)
- Good overview of the natural Xer recombinase system.
| "In strains deleted for dif, failure to resolve dimer chromosomes leads to the Dif phenotype, which includes filamentation in a fraction of the cells, abnormal nucleoid morphology, SOS induction and decreased growth rate and plating efficiency compared with wild-type cells (Kuempel et al., 1991; Cornet et al., 1996)." |
- Summary: Missing or broken dif sites result in filamentation and slower growth rate
| "dif is composed of a 28 bp ‘core’ sequence homologous to cer, which acts as the binding site for the resolvase enzymes XerC and XerD (Blakely et al., 1993). A difference between the sites is that cer is flanked by accessory sequences, which bind additional proteins and enhance recombination between sites in dimers. This provides directionality so that resolution of dimers to monomers is highly favoured (Blakely and Sherratt, 1996). Flanking sequences are not involved in resolution at dif as the phenotype of a 173 kb deletion can be suppressed by insertion of a 33 bp dif sequence (Tecklenburg et al., 1995). It has been proposed that directionality is provided by some aspect of chromosome partitioning itself. Structure is also important, as ectopic dif sites are only fully functional when located in an ≈30 kb region that surrounds the normal location of dif (Cornet et al., 1996; Kuempel et al., 1996)." |
- Summary: cer sites use accessory proteins to enhance dimer->monomer versus the reverse. dif sites don't use accessory proteins and favor the dimer-> monomer direction by some other structural or mechanical mechanism inherent in cell division (possibly KOPS sites and FtsK).
[http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1365-2958.2007.05755.x Bigot]
- a literal chromosome segregation machine (2007)
- An up to date review on the mechanics of FtsK
- Summary: The yellow rings are the FtsK motors which powerfully pull the DNA through them. When they encounter dif sites with XerC and XerD recombinases on them the recombination event is activated
References
Hendricks EC, Szerlong H, Hill T, Kuempel P. Cell division, guillotining of dimer chromosomes and SOS induction in resolution mutants (dif, xerC and xerD) of Escherichia coli. Mol Microbiol. 2000 May;36(4):973-81.
Ptacin JL, Nollmann M, Bustamante C, Cozzarelli NR. Identification of the FtsK sequence-recognition domain. Nat Struct Mol Biol. 2006 Nov;13(11):1023-5. Epub 2006 Oct 15.
Recchia GD, Aroyo M, Wolf D, Blakely G, Sherratt DJ. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 1999 Oct 15;18(20):5724-34.
Steiner WW, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998 Dec;180(23):6269-75.
Strick TR, Quessada-Vial A. FtsK: a groovy helicase. Nat Struct Mol Biol. 2006 Nov;13(11):948-50. Erratum in: Nat Struct Mol Biol. 2007 Jun;14(6):568.