Virginia Tech/plasmid construct
From 2007.igem.org
< Virginia Tech(Difference between revisions)
m |
BlairLyons (Talk | contribs) m |
||
| (One intermediate revision not shown) | |||
| Line 42: | Line 42: | ||
<img src="https://static.igem.org/mediawiki/2007/d/d5/Button_jc.JPG"/></a></html>[[Image:Dot1.JPG]] | <img src="https://static.igem.org/mediawiki/2007/d/d5/Button_jc.JPG"/></a></html>[[Image:Dot1.JPG]] | ||
| - | <!--sixth cell of second table: | + | <!--sixth cell of second table: Contributions and Contact--> |
| style="padding: 0px; width=50px; background-color: #FFFFFF;" | | | style="padding: 0px; width=50px; background-color: #FFFFFF;" | | ||
| - | <html><a href="https://2007.igem.org/Virginia_Tech/ | + | <html><a href="https://2007.igem.org/Virginia_Tech/Contributions"> |
| - | <img src="https://static.igem.org/mediawiki/2007/ | + | <img src="https://static.igem.org/mediawiki/2007/6/69/Button_contribute.JPG"/></a></html><html><a href="mailto:igem@vt.edu"></a></html> |
|}<html></center></html> | |}<html></center></html> | ||
| Line 62: | Line 62: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<!--fourth table: Content--> | <!--fourth table: Content--> | ||
| Line 92: | Line 73: | ||
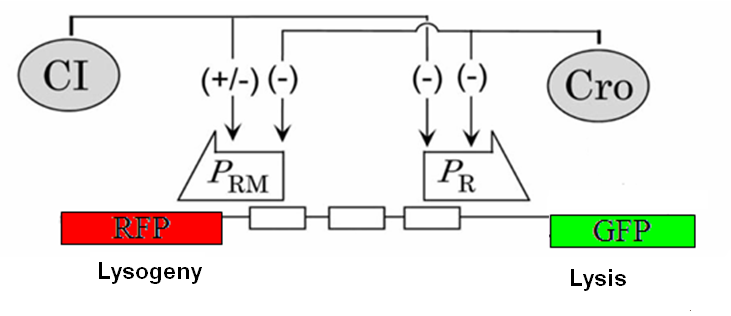
| + | [[Image:Gene_circuit_fl.PNG|thumb|right|400px|'''The reporter plasmid''' the two different fluorescent genes are each on opposite sides of the right promoter of the λ switch.]] | ||
| + | <h3>Our original plan was to build our plasmid in two ways.</h3> We wanted to contrast the iGEM method to more traditional methods of molecular biology. Both approaches required three DNA parts: | ||
| + | |||
| + | 1) A bistable promoter derived from the λ switch (see [[Virginia_Tech/plasmid_design|Designing the Plasmid]]) | ||
| + | |||
| + | 2) Two fluorescent protein genes from the registry | ||
| + | |||
| + | 3) A vector backbone from the registry | ||
| + | While constructing our plasmid, we decided it would be interesting to sequence the registry in order to add more quality control. Other members of our lab undertook this project. More details are [[Virginia_Tech/sequencing|here]]. | ||
| + | Our more traditional plan involved synthesizing the plasmid. We knew this would be a quicker way to get the plasmid so that we could begin testing it and obtain experimental data for our model. The results from this approach did not turn out as we expected. We were delayed in finishing the design and having it synthesized. | ||
|}<html></center></html> | |}<html></center></html> | ||
Latest revision as of 03:24, 27 October 2007
|
|
|
Our original plan was to build our plasmid in two ways.We wanted to contrast the iGEM method to more traditional methods of molecular biology. Both approaches required three DNA parts:1) A bistable promoter derived from the λ switch (see Designing the Plasmid) 2) Two fluorescent protein genes from the registry 3) A vector backbone from the registry
Our more traditional plan involved synthesizing the plasmid. We knew this would be a quicker way to get the plasmid so that we could begin testing it and obtain experimental data for our model. The results from this approach did not turn out as we expected. We were delayed in finishing the design and having it synthesized.
|