Harvard
From 2007.igem.org
(→The Harvard University 2007 iGEM Team) |
(→Bacterial Targeting) |
||
| (86 intermediate revisions not shown) | |||
| Line 32: | Line 32: | ||
=The Harvard University 2007 iGEM Team= | =The Harvard University 2007 iGEM Team= | ||
| - | This year Harvard's team consisted of | + | This year Harvard's team consisted of eight undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, engineering, and computer science. With the help of six faculty advisers and four teaching fellows, plus one education advisor, we devised and executed a single project with three subsections. |
| - | <p style = "font-size:150%;" | + | <p style = "font-size:150%;">Cling-<i>E. Coli</i></p> |
| - | #Bacterial Targeting | + | #[https://2007.igem.org/Harvard#Bacterial_Targeting Bacterial Targeting] |
| - | #Quorum Sensing | + | #[https://2007.igem.org/Harvard#Quorum_Sensing Quorum Sensing] |
| - | # | + | #[https://2007.igem.org/Harvard#Fec_Signal_Transduction Fec Signal Transduction] |
| + | <br> | ||
| + | Our efforts were focused on engineering bacterial surface proteins to express peptides that will target bacteria to specific substrates. We then sought a biological read-out for whether the engineered bacteria interact with the substrate. For this purpose, we employed two distinct detection methods: quorum sensing and the Fec iron response system. | ||
| + | <br> | ||
| + | <br> By the end of the summer, we successfully demonstrated that bacteria expressing well-characterized tags on their surface could bind to specific substrates. Separately, we characterized the quorum system with multiple types of promoters and reporters, and we are on our way toward a combined targeting-quorum system in the near future. In addition, we are working on inserting random peptide libraries into the bacterial surface proteins; we will then screen for the ability to bind to a variety of substrates. | ||
| + | <br> | ||
| + | <br>Eventually, our project may have applicability in medical imaging and localized drug synthesis and release. Theoretically, if our project is successful, bacteria (or other microorganisms) could be targeted specifically to a certain tumor cell. At a certain density, quorum sensing may allow for the bacteria to emit a signal that could be detected by medical imaging devices. If the reporter gene encoded for drugs or vitamins, synthesis can also be evoked through enriched targeting. Furthermore, binding through a modified Fec system may allow for binding alone to transduce a signal and elicit a response in the cells. | ||
| + | ==Bacterial Targeting== | ||
| + | '''Project Overview''' | ||
| + | *The motivation for this part of the project was to engineer bacteria to adhere to targets with a high degree of specificity. | ||
| + | *Initial targeting was done by displaying histidine and strep2 tags on the E. coli surface via fusion with the proteins LppOmpA and AIDA-1, and screens were performed with binding to their known nickel and streptavidin targets, respectively. | ||
| + | *After characterization and high enrichment with these known substrates, random libraries were inserted into LppOmpA and AIDA-1 constructs for screening peptides with affinity for novel targets. As we proceed with this experiment, we hope to characterize sequences that lend specificity to calmodulin and EGF. | ||
| + | *We also tried to utilize the same enrichment strategy with cells that were cotransformed with both AIDA-1-fused tags and quorum-sensing genes (see below). | ||
| - | + | '''Methods''' | |
| + | *Bacteria with various outer-membrane surface expression proteins were engineered his and strep2 tags on the E. coli surface via fusion with LppOmpA and AIDA-1 vehicles: | ||
| + | *#OmpA1 with C-terminus insertion | ||
| + | *#OmpA1 with a Loop1 insertion | ||
| + | *#AIDA-1 with a N-terminus insertion | ||
| + | *In order to test the various ouster-membrane proteins and their ability to bind to specific antibodies and beads (nickel/streptavidin), various cell-sorting assays were run to ascertain the binding strength of the constructs against a background of colonies, along with the robustness of the assays. The following types of assays were run: | ||
| + | *# Magnetic Activated Cell Sorting (MACS) | ||
| + | *#*Mixed Bacterial cultures of cells with strep2 or his tags, along with a background colony with a color (e.g. RFP) distinct from the tagged cells. | ||
| + | *#**In our case we usually used tagged white cells with the AIDA+his or AIDA1+strep2 along with RFP-labeled background cells. | ||
| + | *#*Added his or strep antibodies and magnetic beads and incubated in order to bind to surface expression proteins. | ||
| + | *#*Ran the mixed culture through a magnetic column, so the flow-through would consist of the untagged cells while the bound/tagged cells are stuck with the magnetic bead. After removal of the magnet, we washed the cells out, and plated them to see how much background binding took place. | ||
| + | *# Fluorescence Activated Cell Sorting (FACS) | ||
| + | *#*Similar protocol to the MACS up to the sort step, except for the addition of fluorescent antibodies. | ||
| + | *#*With the two various samples mixed together, the bacteria were taken to the flow cytometry center at Harvard, and we hit the antibodies with a laser of a proper wavelength, and from there we could select for specific cells with various fluorescence. Then the cells were plated to determine whether we had selected the tagged cells over the untagged cells. | ||
| + | '''Results''' | ||
| + | *Successfully engineered his and strep2 tags on both OmpA and AIDA constructs. | ||
| + | *Successfully ran both MACS and FACS assays and received positive results for both types of assays, although we received the best results from the MACS (a more robust system). | ||
| + | *Successfully were able to target bacteria to nickel and streptavidin with our engineered constructs. | ||
| + | <gallery> | ||
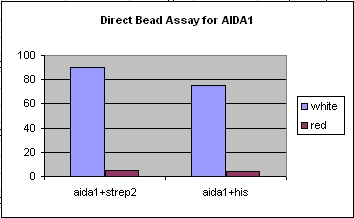
| + | Image:DirectAssay1.jpg|Direct magnetic assay, which does not employ antibodies to bind to the magnetic beads. We selected for the light colored cells against the red color background, receiving positive results for both his and strep. | ||
| + | Image:MACSAssay1.jpg|Indirect magnetic assay, which employs antibodies to bind to the magnetic beads. We selected for the light colored cells against the red color background, receiving positive results for both his and strep. | ||
| + | Image:FACSAssay1.jpg|A type of fluorescence assay, which does not employ antibodies to bind to the magnetic beads. We selected for the light colored cells against the red color background, receiving positive results for the AIDA1+strep. | ||
| + | </gallery> | ||
| + | '''Future Directions ''' | ||
| + | *Trying out new targeting peptides such as calmodulin (CaM, a calcium binding protein) | ||
| + | *Use a random-library approach to select for targeting peptides. | ||
| + | **Ligate random-library 10-mers or 15-mers into plasmid DNA (including potential biobricks) | ||
| + | **Use our engineered membrane proteins to identify surface peptide sequences that target bacteria to various tissue types or cell types we are interested in. | ||
| + | ***This has important medical implications since we may be able to target "microbial factories" to various areas of the body using this method. We can also select for peptides that bind to other cell types. | ||
| + | |||
| + | '''References''' | ||
| + | *''Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines''. Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL, Curtiss R 3rd. Nat Biotechnol. 1997 Jan;15(1):29-34. Review. | ||
| + | |||
| + | *''Autodisplay: efficient bacterial surface display of recombinant proteins.'' Jose J. Appl Microbiol Biotechnol. 2006 Feb;69(6):607-14. Epub 2005 Dec 20. Review. | ||
| + | |||
| + | *''Organ targeting in vivo using phage display peptide libraries''. Pasqualini R, Ruoslahti E. Nature. 1996 Mar 28;380(6572):364-6. | ||
| + | |||
| + | *''Environmentally controlled invasion of cancer cells by engineered bacteria.'' J Mol Biol. 2006 Jan 27;355(4):619-27. Anderson JC, Clarke EJ, Arkin AP, Voigt CA. | ||
| + | |||
| + | *''A bacterial protein enhances the release and efficacy of liposomal cancer drugs.'' Science. 2006 Nov 24;314(5803):1308-11. Cheong I, Huang X, Bettegowda C, Diaz LA Jr, Kinzler KW, Zhou S, Vogelstein B. | ||
==Quorum Sensing== | ==Quorum Sensing== | ||
| + | '''Project Overview''' | ||
| + | *The motivation for this part of the project was to effect downstream activity after E. coli bind to a particular substrate, using the luxI/luxR quorum-sensing system from Vibrio fischeri, which would turn on after the bacteria localize to the target. | ||
| + | *Lux quorum-sensing works like a system of sender and receiver. LuxI codes for a protein that catalyzes the synthesis of 3-oxo-hexanyl homoserine lactone, or OHHL, which can diffuse freely through the surface of E. coli into the environment and into other E. coli. LuxR encodes for a protein which does not diffuse across the membrane and, when bound to OHHL, upregulates the luxpR promoter. So luxI-producing bacteria (senders) produce OHHL that diffuses into luxR-producing bacteria (receivers) where OHHL and luxR turn on luxpR promoter. This only occurs at a certain concentration of OHHL; thus a certain concentration of cells (quorum) is required. | ||
| + | *Initial characterizations of the luxI/luxR system and quorum activity were made using GFP and RFP reporters. Two approaches were taken to quorum sensing. (1) A luxI/luxR production system in one cell would be simpler, but it's possible that the cells might self-induce. (2) luxI and luxR production in separate cells would ensure no self-induction, but it requires monitoring two populations of cells. | ||
| + | |||
| + | '''Methods''' | ||
| + | *To characterize quorum activity, constitutive RFP senders were mixed with constitutive receivers. Fluorescence and OD (cell density) readings were taken every 15 minutes. [http://openwetware.org/wiki/IGEM:Harvard/2007/Laboratory_Notebooks/Quorum_Sensing/Plate_Reader_Protocol The protocol] was adjusted as necessary according to what the exact desired experiment was. | ||
| + | *To characterize binding activity, RFP-senders with AIDA tags were mixed with constitutive GFP non-tagged cells. As a control, a portion of the mixed cells was plated pre-selection. Direct magnetic beads (TALON nickel, Qiagen streptactin) were used to enrich for the tagged-senders. Several washes were done in low concentration imidazole. After incubations with beads and washes, the mixture was plated and grown overnight, and cell counts were done to determine extent of enrichment. (A more detailed protocol [http://openwetware.org/wiki/Shlo/directmagneticprotocol available]). | ||
| + | *More detailed methods are available on our [http://openwetware.org/wiki/IGEM:Harvard/2007/Laboratory_Notebooks/Quorum_Sensing wiki]. | ||
| + | |||
| + | '''Results''' | ||
| + | *We were able to demonstrate that the sender-receiver system in one cell was not self-inducing but acted with quorum activity. | ||
| + | *Quorum activity was also detected with the separate sender and receiver cells. | ||
| + | *In collaboration with the [http://openwetware.org/wiki/IGEM:Harvard/2007/Bacterial_Targeting bacterial targeting] subteam, we have evidence that red sender cells tagged with strep2 on the surface (cotransformed with constitutive RFP/luxI plasmid and AIDA-strep2 plasmid) are enriched by selection with streptavidin-coated beads, while green non-tagged cells are not. | ||
| + | *Quorum activity was demonstrated as well through a plate-drop experiment. | ||
| + | *[http://openwetware.org/wiki/Shlo/flmicroprotstickiness Microscopy] was also used to detect and image tagged bacteria accumulating around beads, while non-tagged bacteria did not aggregate. | ||
| + | <gallery> | ||
| + | Image:Harvigem07directmag.jpg|Enrichment by selection with magnetic beads | ||
| + | Image:Harvigem07quorumcurve.jpg|Fluorescence data indicates quorum activity | ||
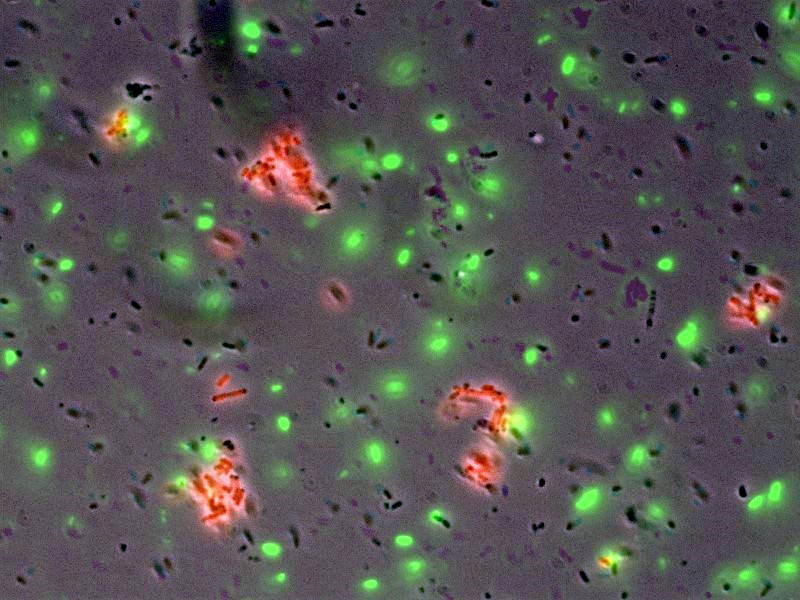
| + | Image:Harvigem07microscopyt.jpg|Microscopy images show tagged, red-sender cells accumulate around the beads, whereas the background green cells did not | ||
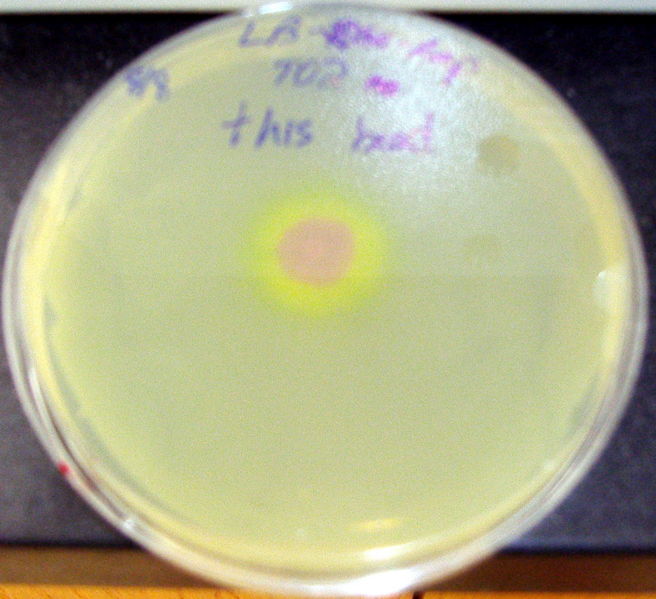
| + | Image:Harvarigem07platequorum.jpg|Quorum activity was demonstrated via magnetically enriched sender cells that were plated on a receiver plate. | ||
| + | </gallery> | ||
| + | '''Conclusions and future plans''' | ||
| + | *Continue characterization of single cell constructs (approach (1) under Project Overview). | ||
| + | *Characterize cell and bead density require to reach quorum levels. We have been attempting to do this through Nickel-coated 96 well plates. | ||
| + | |||
| + | '''References''' | ||
| + | *Voigt CA. Genetic parts to program bacteria. Curr Opin Biotechnol 2006 Oct; 17(5) 548-57. doi:10.1016/j.copbio.2006.09.001 pmid:16978856. | ||
| + | *Anderson JC, Clarke EJ, Arkin AP, and Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 2006 Jan 27; 355(4) 619-27. doi:10.1016/j.jmb.2005.10.076 pmid:16330045 | ||
| + | *Basu S, Mehreja R, Thiberge S, Chen MT, and Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004 Apr 27;101(17):6355-60. | ||
| + | *Sayut DJ, Niu Y, Sun L. Construction and engineering of positive feedback loops. ACS Chem Biol. 2006 Dec 20;1(11):681-2. | ||
| + | |||
| + | ==Fec Signal Transduction== | ||
| + | |||
| + | '''Project Overview''' | ||
| + | *The motivation for this project was to create a system of targetting and direct signalling. In other words, E.Coli would bind to a target ligand, causing signal transduction and gene expression. This gene expression would cause a relevant reaction in the presence of the ligand. In a "Save the World" scenario, E.Coli could bind to a surface protein common in cancer cells, and then express cytotoxins to kill the cells. <br> | ||
| + | |||
| + | *We began by searching for known E.Coli signalling systems. The Fec system was chosen because it is the only known and well characterized signalling system with an outermembrane receptor. Most other signalling systems respond to substrates that diffuse into the periplasm and bind to inner membrane proteins. This would be a very limiting mechanism for targetting, as the outer membrane would need to be permeable to any chosen target.<br> | ||
| + | *Fec System Overview: | ||
| + | **The Fec system receptor is the outer membrane protein FecA. Its wild-type ligand is dinuclear ferric citrate. | ||
| + | **When binding occurs in iron-limiting conditions, a signal is sent to the inner membrane protein FecR. | ||
| + | **FecR activates sigma factor FecI. | ||
| + | **FecI is then used to induce gene expression.<br> | ||
| + | *Structural papers detail the conformational changes that FecA undergoes when binding to ferric citrate. | ||
| + | **The alpha helix in loop 7 unravels, and the loop moves by up to 11 angstroms | ||
| + | **Loop 8 moves up to 15 angstroms. | ||
| + | *The motion of these two loops closes over the ferric citrate.<br> | ||
| + | *These large changes suggest the importance of loops 7 and 8 to binding. From this, we proposed three options to modify the binding of FecA and hopefully maintain signalling:<br> | ||
| + | **Inserting a binding sequence into loop 7 (for simplicity and time, we arbitrarily chose to use either 7 or 8, and ended up using loop 7) of FecA such that it would bind to a new target at least, and produce a signal at most. | ||
| + | **A computational approach which would produce nucleotide sequences for binding our new target. A directed mutagenesis of FecA would follow. | ||
| + | ** Use of random libraries to find binding sequences for given targets. | ||
| + | '''Methods'''<br> | ||
| + | *Our first approach was to insert binding sequences into FecA. We chose 6xHis and Strep as binding sequences for nickel and streptavidin, respectively. | ||
| + | *The main constructs used were shipped from researcher Volkmar Braun who studied the Fec system at the University of Tuebingen in Germany. The constructs obtained were:<br> | ||
| + | **1) AA93 E.Coli strain which does not express the Fec system | ||
| + | **2) A GFP plasmid with a Fec promotor and, | ||
| + | **3) pLCIRA, a plasmid containing the Fec system genes.<br> | ||
| + | *These constructs were necessary because in order to test the binding of our re-engineered FecA, we would need to find a system or strain of E.Coli such that the natural Fec genes would be knocked out and only our re-engineering FecA would be expressed. These part constitute the first system used for binding site insertion.<br> | ||
| - | + | *Because pLCIRA is not well characterized and the expression of the Fec system is under its own promoter and upregulates itself upon binding, we thought it valuable to be able to control levels of FecA expression. So, we attempted to use a second system. This second system still included the GFP plasmid from system 1, but we replaced the pLCIRA with a T7 pColA duet vector. This way, expression was induced and control by the addition of IPTG. However, AA93 is not a T7 cell, so we used lysogenization to make AA93 DE3 cells.<br> | |
| + | *We then inserted FecI&R (pcr'ed from the pLCIRA) into the first cloning site of the pColA plasmid and FecA (also pcr'ed from pLCIRA) into the second.<br> | ||
| + | See the Harvard iGEM 2007 wiki for more [http://openwetware.org/wiki/IGEM:Harvard/2007/Laboratory_Notebooks/Two_Component_System details]... | ||
| + | '''Results''' | ||
| + | *The pColA system was difficult to work with. Our cells had trouble surviving, especially cultures that had leaky Fec system expression. We believe that overexpression of the Fec system killed the cells, possibly by disturbing the cell membranes. The cells inconclusive fluorescence in the presence of the wild type ligand. To explain the assay and results: | ||
| + | **We used sodium citrate, not ferric citrate, to induce. We believe this works because the citrate chelated free iron out of the LB, thus forming ferric citrate. Ferric citrate, however, does not deal with the free iron and the Fec system is repressed by the Fur repressor in the presence of free iron. | ||
| + | **Our assay with the pColA system showed fluorescence above the uninduced(no sodium citrate) levels, but very little (130 RFUs at most). And there is no clear trend of IPTG vs no-IPTG or for the different concentrations of sodium citrate. These results are expressed in the first graph below. | ||
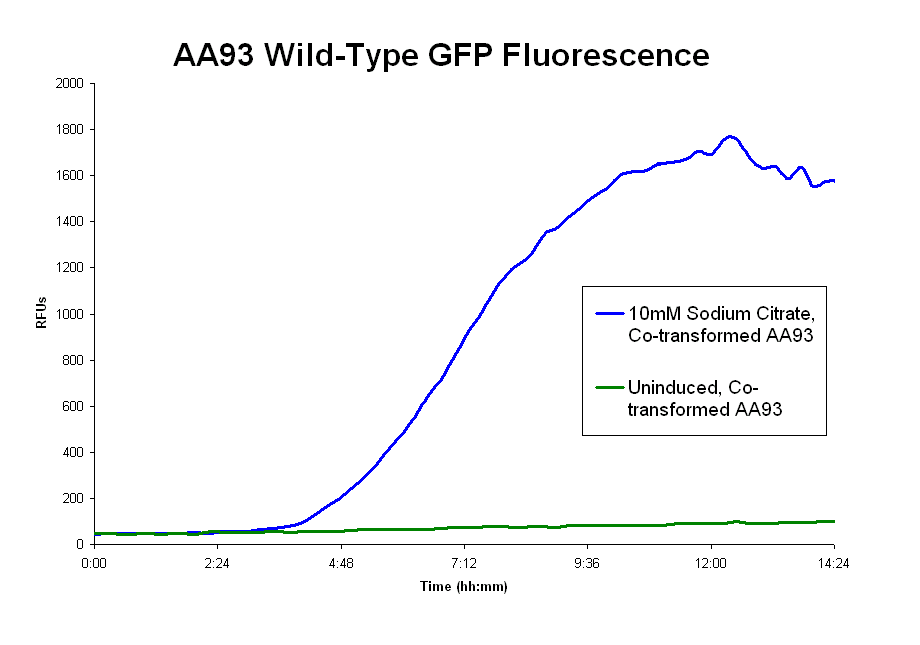
| + | *The pLCIRA system is still in the works. We were able to get fluorescence with the wild type ligand (second graph below), though even this took much effort. We tried out different concentrations and 10mM sodium citrate worked best. The strep and 6xHis binding sequences have been added to FecA, but testing has yet to occur. No results as yet. | ||
| + | '''Conclusions and future plans''' | ||
| + | *We have yet to prove that the Fec system is viable for targetting. We are currently working with the pLCIRA system and will hopefully test for binding in the coming week(s). | ||
| + | <gallery> | ||
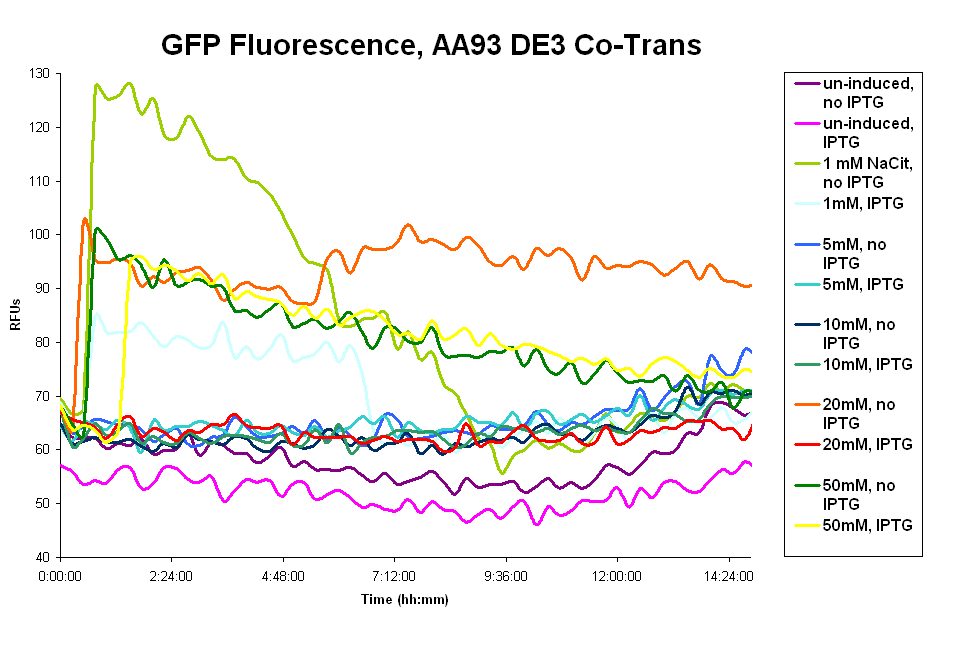
| + | Image:GFP_de3_fluorescence.png|GFP fluorescence data of Co-transformed de3 AA93 cells. | ||
| + | Image:GFP_fluorescence.png|GFP fluorescence data of Co-transformed AA93 with wildtype ligand | ||
| + | </gallery> | ||
| + | '''References''' | ||
| + | *Braun V, Mahren S, and Sauter A. Gene regulation by transmembrane signaling. Biometals 2006 Apr; 19(2) 103-13. | ||
| + | *Ferguson AD, Amezcua CA, Halabi NM, Chelliah Y, Rosen MK, Ranganathan R, and Deisenhofer J. Signal transduction pathway of TonB-dependent transporters. Proc Natl Acad Sci U S A 2007 Jan 9; 104(2) 513-8 | ||
| + | *Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, and Deisenhofer J. Structural basis of gating by the outer membrane transporter FecA. Science 2002 Mar 1; 295(5560) 1715-9. | ||
| + | *Sauter A and Braun V. Defined inactive FecA derivatives mutated in functional domains of the outer membrane transport and signaling protein of Escherichia coli K-12. J Bacteriol 2004 Aug; 186(16) 5303-10. | ||
| + | *[http://openwetware.org/wiki/IGEM:Harvard/2007/Two_Component_Systems Further reading] on Fec system (scroll down to "Readings").<br> | ||
<html><a href="http://openwetware.org/wiki/IGEM:Harvard/2007"><img src="http://openwetware.org/images/4/42/ThumbWiki.png"></a></html><br> | <html><a href="http://openwetware.org/wiki/IGEM:Harvard/2007"><img src="http://openwetware.org/images/4/42/ThumbWiki.png"></a></html><br> | ||
<p style = "font-size:120%;">Click the picture, or click [http://openwetware.org/wiki/IGEM:Harvard here] for the <b>more detailed</b> Harvard iGEM 2007 wiki.</p> | <p style = "font-size:120%;">Click the picture, or click [http://openwetware.org/wiki/IGEM:Harvard here] for the <b>more detailed</b> Harvard iGEM 2007 wiki.</p> | ||
Latest revision as of 03:36, 27 October 2007
| Harvard University 2007 iGEM Team | ||
Students
Education Advisor
|
Teaching Fellows
Faculty Advisors
| |
The Harvard University 2007 iGEM Team
This year Harvard's team consisted of eight undergraduate students, with backgrounds in molecular and cellular biology, biochemistry, engineering, and computer science. With the help of six faculty advisers and four teaching fellows, plus one education advisor, we devised and executed a single project with three subsections.
Cling-E. Coli
Our efforts were focused on engineering bacterial surface proteins to express peptides that will target bacteria to specific substrates. We then sought a biological read-out for whether the engineered bacteria interact with the substrate. For this purpose, we employed two distinct detection methods: quorum sensing and the Fec iron response system.
By the end of the summer, we successfully demonstrated that bacteria expressing well-characterized tags on their surface could bind to specific substrates. Separately, we characterized the quorum system with multiple types of promoters and reporters, and we are on our way toward a combined targeting-quorum system in the near future. In addition, we are working on inserting random peptide libraries into the bacterial surface proteins; we will then screen for the ability to bind to a variety of substrates.
Eventually, our project may have applicability in medical imaging and localized drug synthesis and release. Theoretically, if our project is successful, bacteria (or other microorganisms) could be targeted specifically to a certain tumor cell. At a certain density, quorum sensing may allow for the bacteria to emit a signal that could be detected by medical imaging devices. If the reporter gene encoded for drugs or vitamins, synthesis can also be evoked through enriched targeting. Furthermore, binding through a modified Fec system may allow for binding alone to transduce a signal and elicit a response in the cells.
Bacterial Targeting
Project Overview
- The motivation for this part of the project was to engineer bacteria to adhere to targets with a high degree of specificity.
- Initial targeting was done by displaying histidine and strep2 tags on the E. coli surface via fusion with the proteins LppOmpA and AIDA-1, and screens were performed with binding to their known nickel and streptavidin targets, respectively.
- After characterization and high enrichment with these known substrates, random libraries were inserted into LppOmpA and AIDA-1 constructs for screening peptides with affinity for novel targets. As we proceed with this experiment, we hope to characterize sequences that lend specificity to calmodulin and EGF.
- We also tried to utilize the same enrichment strategy with cells that were cotransformed with both AIDA-1-fused tags and quorum-sensing genes (see below).
Methods
- Bacteria with various outer-membrane surface expression proteins were engineered his and strep2 tags on the E. coli surface via fusion with LppOmpA and AIDA-1 vehicles:
- OmpA1 with C-terminus insertion
- OmpA1 with a Loop1 insertion
- AIDA-1 with a N-terminus insertion
- In order to test the various ouster-membrane proteins and their ability to bind to specific antibodies and beads (nickel/streptavidin), various cell-sorting assays were run to ascertain the binding strength of the constructs against a background of colonies, along with the robustness of the assays. The following types of assays were run:
- Magnetic Activated Cell Sorting (MACS)
- Mixed Bacterial cultures of cells with strep2 or his tags, along with a background colony with a color (e.g. RFP) distinct from the tagged cells.
- In our case we usually used tagged white cells with the AIDA+his or AIDA1+strep2 along with RFP-labeled background cells.
- Added his or strep antibodies and magnetic beads and incubated in order to bind to surface expression proteins.
- Ran the mixed culture through a magnetic column, so the flow-through would consist of the untagged cells while the bound/tagged cells are stuck with the magnetic bead. After removal of the magnet, we washed the cells out, and plated them to see how much background binding took place.
- Mixed Bacterial cultures of cells with strep2 or his tags, along with a background colony with a color (e.g. RFP) distinct from the tagged cells.
- Fluorescence Activated Cell Sorting (FACS)
- Similar protocol to the MACS up to the sort step, except for the addition of fluorescent antibodies.
- With the two various samples mixed together, the bacteria were taken to the flow cytometry center at Harvard, and we hit the antibodies with a laser of a proper wavelength, and from there we could select for specific cells with various fluorescence. Then the cells were plated to determine whether we had selected the tagged cells over the untagged cells.
- Magnetic Activated Cell Sorting (MACS)
Results
- Successfully engineered his and strep2 tags on both OmpA and AIDA constructs.
- Successfully ran both MACS and FACS assays and received positive results for both types of assays, although we received the best results from the MACS (a more robust system).
- Successfully were able to target bacteria to nickel and streptavidin with our engineered constructs.
Future Directions
- Trying out new targeting peptides such as calmodulin (CaM, a calcium binding protein)
- Use a random-library approach to select for targeting peptides.
- Ligate random-library 10-mers or 15-mers into plasmid DNA (including potential biobricks)
- Use our engineered membrane proteins to identify surface peptide sequences that target bacteria to various tissue types or cell types we are interested in.
- This has important medical implications since we may be able to target "microbial factories" to various areas of the body using this method. We can also select for peptides that bind to other cell types.
References
- Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Georgiou G, Stathopoulos C, Daugherty PS, Nayak AR, Iverson BL, Curtiss R 3rd. Nat Biotechnol. 1997 Jan;15(1):29-34. Review.
- Autodisplay: efficient bacterial surface display of recombinant proteins. Jose J. Appl Microbiol Biotechnol. 2006 Feb;69(6):607-14. Epub 2005 Dec 20. Review.
- Organ targeting in vivo using phage display peptide libraries. Pasqualini R, Ruoslahti E. Nature. 1996 Mar 28;380(6572):364-6.
- Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006 Jan 27;355(4):619-27. Anderson JC, Clarke EJ, Arkin AP, Voigt CA.
- A bacterial protein enhances the release and efficacy of liposomal cancer drugs. Science. 2006 Nov 24;314(5803):1308-11. Cheong I, Huang X, Bettegowda C, Diaz LA Jr, Kinzler KW, Zhou S, Vogelstein B.
Quorum Sensing
Project Overview
- The motivation for this part of the project was to effect downstream activity after E. coli bind to a particular substrate, using the luxI/luxR quorum-sensing system from Vibrio fischeri, which would turn on after the bacteria localize to the target.
- Lux quorum-sensing works like a system of sender and receiver. LuxI codes for a protein that catalyzes the synthesis of 3-oxo-hexanyl homoserine lactone, or OHHL, which can diffuse freely through the surface of E. coli into the environment and into other E. coli. LuxR encodes for a protein which does not diffuse across the membrane and, when bound to OHHL, upregulates the luxpR promoter. So luxI-producing bacteria (senders) produce OHHL that diffuses into luxR-producing bacteria (receivers) where OHHL and luxR turn on luxpR promoter. This only occurs at a certain concentration of OHHL; thus a certain concentration of cells (quorum) is required.
- Initial characterizations of the luxI/luxR system and quorum activity were made using GFP and RFP reporters. Two approaches were taken to quorum sensing. (1) A luxI/luxR production system in one cell would be simpler, but it's possible that the cells might self-induce. (2) luxI and luxR production in separate cells would ensure no self-induction, but it requires monitoring two populations of cells.
Methods
- To characterize quorum activity, constitutive RFP senders were mixed with constitutive receivers. Fluorescence and OD (cell density) readings were taken every 15 minutes. [http://openwetware.org/wiki/IGEM:Harvard/2007/Laboratory_Notebooks/Quorum_Sensing/Plate_Reader_Protocol The protocol] was adjusted as necessary according to what the exact desired experiment was.
- To characterize binding activity, RFP-senders with AIDA tags were mixed with constitutive GFP non-tagged cells. As a control, a portion of the mixed cells was plated pre-selection. Direct magnetic beads (TALON nickel, Qiagen streptactin) were used to enrich for the tagged-senders. Several washes were done in low concentration imidazole. After incubations with beads and washes, the mixture was plated and grown overnight, and cell counts were done to determine extent of enrichment. (A more detailed protocol [http://openwetware.org/wiki/Shlo/directmagneticprotocol available]).
- More detailed methods are available on our [http://openwetware.org/wiki/IGEM:Harvard/2007/Laboratory_Notebooks/Quorum_Sensing wiki].
Results
- We were able to demonstrate that the sender-receiver system in one cell was not self-inducing but acted with quorum activity.
- Quorum activity was also detected with the separate sender and receiver cells.
- In collaboration with the [http://openwetware.org/wiki/IGEM:Harvard/2007/Bacterial_Targeting bacterial targeting] subteam, we have evidence that red sender cells tagged with strep2 on the surface (cotransformed with constitutive RFP/luxI plasmid and AIDA-strep2 plasmid) are enriched by selection with streptavidin-coated beads, while green non-tagged cells are not.
- Quorum activity was demonstrated as well through a plate-drop experiment.
- [http://openwetware.org/wiki/Shlo/flmicroprotstickiness Microscopy] was also used to detect and image tagged bacteria accumulating around beads, while non-tagged bacteria did not aggregate.
Conclusions and future plans
- Continue characterization of single cell constructs (approach (1) under Project Overview).
- Characterize cell and bead density require to reach quorum levels. We have been attempting to do this through Nickel-coated 96 well plates.
References
- Voigt CA. Genetic parts to program bacteria. Curr Opin Biotechnol 2006 Oct; 17(5) 548-57. doi:10.1016/j.copbio.2006.09.001 pmid:16978856.
- Anderson JC, Clarke EJ, Arkin AP, and Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 2006 Jan 27; 355(4) 619-27. doi:10.1016/j.jmb.2005.10.076 pmid:16330045
- Basu S, Mehreja R, Thiberge S, Chen MT, and Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004 Apr 27;101(17):6355-60.
- Sayut DJ, Niu Y, Sun L. Construction and engineering of positive feedback loops. ACS Chem Biol. 2006 Dec 20;1(11):681-2.
Fec Signal Transduction
Project Overview
- The motivation for this project was to create a system of targetting and direct signalling. In other words, E.Coli would bind to a target ligand, causing signal transduction and gene expression. This gene expression would cause a relevant reaction in the presence of the ligand. In a "Save the World" scenario, E.Coli could bind to a surface protein common in cancer cells, and then express cytotoxins to kill the cells.
- We began by searching for known E.Coli signalling systems. The Fec system was chosen because it is the only known and well characterized signalling system with an outermembrane receptor. Most other signalling systems respond to substrates that diffuse into the periplasm and bind to inner membrane proteins. This would be a very limiting mechanism for targetting, as the outer membrane would need to be permeable to any chosen target.
- Fec System Overview:
- The Fec system receptor is the outer membrane protein FecA. Its wild-type ligand is dinuclear ferric citrate.
- When binding occurs in iron-limiting conditions, a signal is sent to the inner membrane protein FecR.
- FecR activates sigma factor FecI.
- FecI is then used to induce gene expression.
- Structural papers detail the conformational changes that FecA undergoes when binding to ferric citrate.
- The alpha helix in loop 7 unravels, and the loop moves by up to 11 angstroms
- Loop 8 moves up to 15 angstroms.
- The motion of these two loops closes over the ferric citrate.
- These large changes suggest the importance of loops 7 and 8 to binding. From this, we proposed three options to modify the binding of FecA and hopefully maintain signalling:
- Inserting a binding sequence into loop 7 (for simplicity and time, we arbitrarily chose to use either 7 or 8, and ended up using loop 7) of FecA such that it would bind to a new target at least, and produce a signal at most.
- A computational approach which would produce nucleotide sequences for binding our new target. A directed mutagenesis of FecA would follow.
- Use of random libraries to find binding sequences for given targets.
Methods
- Our first approach was to insert binding sequences into FecA. We chose 6xHis and Strep as binding sequences for nickel and streptavidin, respectively.
- The main constructs used were shipped from researcher Volkmar Braun who studied the Fec system at the University of Tuebingen in Germany. The constructs obtained were:
- 1) AA93 E.Coli strain which does not express the Fec system
- 2) A GFP plasmid with a Fec promotor and,
- 3) pLCIRA, a plasmid containing the Fec system genes.
- These constructs were necessary because in order to test the binding of our re-engineered FecA, we would need to find a system or strain of E.Coli such that the natural Fec genes would be knocked out and only our re-engineering FecA would be expressed. These part constitute the first system used for binding site insertion.
- Because pLCIRA is not well characterized and the expression of the Fec system is under its own promoter and upregulates itself upon binding, we thought it valuable to be able to control levels of FecA expression. So, we attempted to use a second system. This second system still included the GFP plasmid from system 1, but we replaced the pLCIRA with a T7 pColA duet vector. This way, expression was induced and control by the addition of IPTG. However, AA93 is not a T7 cell, so we used lysogenization to make AA93 DE3 cells.
- We then inserted FecI&R (pcr'ed from the pLCIRA) into the first cloning site of the pColA plasmid and FecA (also pcr'ed from pLCIRA) into the second.
See the Harvard iGEM 2007 wiki for more [http://openwetware.org/wiki/IGEM:Harvard/2007/Laboratory_Notebooks/Two_Component_System details]...
Results
- The pColA system was difficult to work with. Our cells had trouble surviving, especially cultures that had leaky Fec system expression. We believe that overexpression of the Fec system killed the cells, possibly by disturbing the cell membranes. The cells inconclusive fluorescence in the presence of the wild type ligand. To explain the assay and results:
- We used sodium citrate, not ferric citrate, to induce. We believe this works because the citrate chelated free iron out of the LB, thus forming ferric citrate. Ferric citrate, however, does not deal with the free iron and the Fec system is repressed by the Fur repressor in the presence of free iron.
- Our assay with the pColA system showed fluorescence above the uninduced(no sodium citrate) levels, but very little (130 RFUs at most). And there is no clear trend of IPTG vs no-IPTG or for the different concentrations of sodium citrate. These results are expressed in the first graph below.
- The pLCIRA system is still in the works. We were able to get fluorescence with the wild type ligand (second graph below), though even this took much effort. We tried out different concentrations and 10mM sodium citrate worked best. The strep and 6xHis binding sequences have been added to FecA, but testing has yet to occur. No results as yet.
Conclusions and future plans
- We have yet to prove that the Fec system is viable for targetting. We are currently working with the pLCIRA system and will hopefully test for binding in the coming week(s).
References
- Braun V, Mahren S, and Sauter A. Gene regulation by transmembrane signaling. Biometals 2006 Apr; 19(2) 103-13.
- Ferguson AD, Amezcua CA, Halabi NM, Chelliah Y, Rosen MK, Ranganathan R, and Deisenhofer J. Signal transduction pathway of TonB-dependent transporters. Proc Natl Acad Sci U S A 2007 Jan 9; 104(2) 513-8
- Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, and Deisenhofer J. Structural basis of gating by the outer membrane transporter FecA. Science 2002 Mar 1; 295(5560) 1715-9.
- Sauter A and Braun V. Defined inactive FecA derivatives mutated in functional domains of the outer membrane transport and signaling protein of Escherichia coli K-12. J Bacteriol 2004 Aug; 186(16) 5303-10.
- [http://openwetware.org/wiki/IGEM:Harvard/2007/Two_Component_Systems Further reading] on Fec system (scroll down to "Readings").
Click the picture, or click [http://openwetware.org/wiki/IGEM:Harvard here] for the more detailed Harvard iGEM 2007 wiki.